DNA mutations and repair
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
What different types of DNA mutations are there?
Mutations are changes to DNA. There are several different types;
Point mutations - change, insert or delete a single nucleotide. In regards to a change in nucleotide, these can be transitions or transversions. The former refers to a change in the same type of base (pyrimidine - T, C, purine - A, G) and the latter is the changes between different types of bases. Each of these do not have equal chances of occurring, there is a bias.
Large-scale deletions or insertions.
Repetitive sequences ‘hick-ups’ - these occur when there are small repetitive sequences, and DNA polymerase either adds more or loses some repetitions. These occur at a rapid rate so can be used for forensics - known as microsatellites.
Chromosomal rearrangement - when a chunk of one chromosome replaces another, or swap places.
What are the sources for small-scale mutations?
There are 2 sources for small-scale mutations (point) in DNA. The inaccuracy of DNA replication machinery, and damage to DNA - this can occur through various chemical reactions such as metabolism, radiation, and environmental chemicals.
What are the different severities of mutations?
Mutations can be important - evolution, diversity ect. Some mutations are beneficial, many are neutral/silent, and most are deleterious if not deadly.
Silent mutations are a mutation where a change in base pair does not result in a change in the overall amino acid sequence of a protein. This is due to redundancy.
Missense mutations do result in a change in the amino acid sequence. These can be either good, neutral or bad.
Nonsense mutations are when a premature stop codon is made so that the full protein is not made. This is almost always bad.
How often does DNA polymerase make a mistake and how does it correct this?
DNA polymerase (isolated) makes a mistake for every 10^5 copied base pairs, without any other DNA mechanism. There are 2 ways in which a cell corrects these mistakes; proofreading or mismatch repair.
How does proofreading occur?
When DNA polymerase makes a mistake, it results in a ‘kink’ of the sequence which it detects. It then recruits exonuclease which removes the last added base to the 3’ end. DNA polymerase then continues. This is proofreading, and decreases the error to 1 in every 10^7.
How does mismatch repair occur?
Mismatch repair corrects what escapes proofreading before permanent damage is done (before the cell divides). This means it must scan the genome, find the mutation and fix it quickly.
MutS is a dimer that scans the DNA to find kinks. It then makes the kink more pronounced using energy. It then recruits MutL which activates MutH (always present on the DNA). MutH causes an excision (kink) on the strand near the mismatch site/kink.
Helicase then comes and unwinds the DNA from the excision to the site so that exonuclease can remove DNA from the strand opposite to MutS (this happens as all the other DNA is covered), including the mismatch. DNA polymerase then fills the gap, and ligase attaches the DNA.
How does MutH know which strand to nick?
Dam methylase methylates DNA at every 5’-GATC-3’. After semi-conservative replication, each DNA molecule is only hemi-methylated (for some time) - one strand has methyl groups. The period in which the other strand is not methylated is when mismatch repair occurs.
MutH always sits on the methylated and nicks only the non-methylated strand, which is how MutH always knows which strand to nick/has the mutation.
What is mismatch repair like in eukaryotes?
The MutS/MutL/MutH system is found in E. coli. In eukaryotes, it is done by MSH and MLH, but there is no homologue of MutH.
How does spontaneous damage to DNA occur?
Spontaneous DNA damage occurs because most chemical reactions in biology are reversible. They happen without outside influence. In particular, deamination and hydrolysis;
Deamination of cytosine makes uracil via the addition of water.
Depurination of guanine makes apurinic deoxyribose via addition of water. This removes the guanine base from the sugar-phosphate backbone. This can occur with other bases to make abasic sugars.
Deamination of 5-methylcytosine makes thymine via addition of water.
How does environmental damage occur to DNA?
Environmental damage to DNA can occur mainly through alkylation, oxidation and radiation.
Alkylation is the addition of an alkyl (methyl, ethyl) group to reactive sites on bases or to phosphates in the DNA backbone. For example, the methylation of oxygen 6 in guanine produces O6-methylguanine which pairs with thymine.
Oxidation is the addition of reactive oxygen species (free radicals etc - generated in metabolism), generated by ionising radiation or by chemical agents. The oxidation of guanine produces 8-oxoG which pairs with adenine - this is the most common mutation in cancer.
UV radiation of 260 nm wavelength is particularly dangerous as it leads to the fusion of 2 pyrimidines. This prevents normal base pairing and severely disrupts the local structure of DNA - stops polymerase during replication. Gamma and X-rays cause double stranded breaks, which are difficult to repair as there is no template strand/reference.
What is the Ames test and how does it work?
The Ames test is used to see whether a substance is carcinogenic. First, an engineered salmonella (cannot synthesise histidine) is grown with rat liver extract to simulate the conditions within a normal human. Only a single nonsense mutation is added (STOP codon) so that the His+ gene becomes His-. Because there is only one, a mutation can occur in the same location, causing a reversal.
For a control, a salmonella culture is grown on a plate with minimal histidine (so cannot grow) and is left.
For the test, the salmonella culture is mixed with a potential carcinogen and then grown with minimal histidine. If we see a high number of colonies, more salmonella have mutated from His- to His+.
What DNA repair mechanisms are there?
There are several types of DNA damage repair; direct reversal of DNA damage (fixing damage arising from spontaneous damage), nucleotide/base excision repair, and large-scale damage repair.
What are the methods of direct repair?
For direct reversal -
Removal of a methyl group after alkylation is done by methyltransferase. The kink in the DNa caused by the damage is detected by the enzyme. It then uses its SH group to take off the CH3 group from the DNA in place of its proton.
Photoreactivation reverses the dimerisation done by UV. This is carried out by DNA photolyase. This enzyme requires darkness to detect the dimers, and binds to them. Exposure to visible light induces a reaction to separate the dimer.
How does base excision repair occur?
Base excision repair is one of the most important repair mechanisms. Firstly, a lesion is identified - a damaged base pair in the DNA backbone. Glycosylase recognises the lesions and then removes the base via cleavage of the glycosidic bond between the base and pentose sugar. This leaves behind an abasic sugar. An endo/exonuclease will then cut both ends of the sugar to remove it, then polymerase synthesises the correct sugar, then ligase connects it.
What different glycosylases are there?
There are different glycosylases which each recognise a different lesion;
Uracil glycosylase - detects when cysteine is damaged to become uracil.
8-oxoG glycosylase - detects when guanine is damaged.
3-methyladenine glycosylase - detects when adenine is damaged.
Where do glycosylases struggle?
Glycosylases do not recognise what the original strand was - like with the DNA methylation mechanism (parent vs daughter) - therefore they struggle with mutations that result in a normal DNA base, but wrong pairing.
For example, deamination of methylated cysteine results in a G:T (since it becomes thymine) pairing which is unable to be detected by normal glycosylases as they do not know whether G or T is correct. This requires a specialised glycosylase that identifies this particular pairing and ALWAYS changes the T to a C - this is driven by evolution as this mutation is more common.
How does nucleotide excision repair occur?
Nucleotide excision repair recognises distortions in DNA - these can be caused by a thymine dimer, or a chemical adduct to a base. Detection of distortion is followed by the removal of a short single stranded segment which is then filled up, using the undamaged stand as a template.
UvrA scans the DNA to find distortions. At this point, it is bound to UvrB to form a UvrAB complex - B does nothing right now.
Once a distortion is detected, UvrA leaves the complex. UvrB then melts the DNA to create a single stranded bubble around the distortion which allows access.
UvrB then recruits UvrC which cuts DNA upstream and downstream of the distortion.
This then recruits UvrD, a DNA helicase, which releases the single stranded segment from the duplex by hydrolysing the hydrogen bonds.
DNA polymerase synthesises a new strand, DNA ligase joins it to the DNA molecule.
UvrA, B, C and D are specific to E. coli. In eukaryotes, similar systems exist but include around 25 polypeptides.
How does transcription-coupled repair occur?
Distortions in the DNA slow down or stop RNA polymerase during transcription - detection. This allows transcription-coupled repair to take place, whereby RNA polymerase backs up and recruits UvrAB after encountering a distortion. Nucleotide excision repair occurs whilst RNA polymerase is still attached.
How does double stranded break repair occur?
Double-stranded break repair replies on the fact that there are sister chromatids with the same DNA. It uses homologous recombination to fix this. Firstly, the ends are resections so that one strand overhangs the other (only one strand is cut). These overhangs enable strand invasion of the sister chromatid, so that the non-damaged stand can serve as templates for the damaged strands. DNA polymerase extends the damaged strand, and DNA ligase joins it creating Holliday junctions (marked in yellow arrows) - which get resolved to give rise to repaired DNA. This only works in eukaryotes as we have sister chromatids.
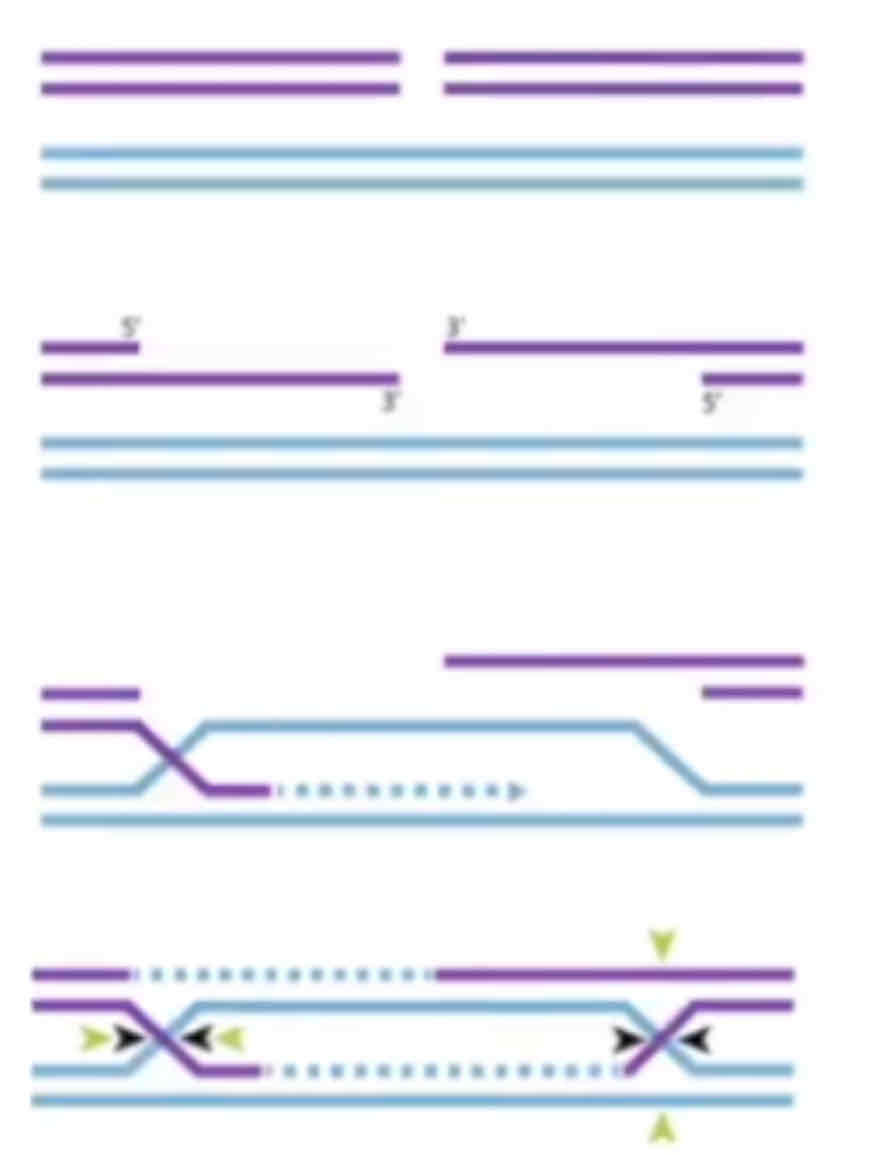
What is non-homologous end joining?
Non-homologous end joining is done by both eukaryotes and prokaryotes. It is a fail safe mechanism by which the ends of chromosomes are stuck together using DNA ligase. This results in potential large mistakes permanently encoded in the DNA, but this is better than a break.
What is translesion and how does it occur?
Some types of damage can disrupt DNA polymerase, but there are no mechanisms to fix this. When DNA polymerase stalls, DNA would not be fully replicated to amplify the damage. Translesion allows DNA polymerase to skip the damage.
DNA polymerase stalls, separates and then Y-polymerase is recruited.
Y-polymerases are a specialised family that incorporate base pairs independently of base pairing of the other strand. Once it passes the damage, DNA polymerase is re-recruited.
This process is very error-prone, therefore cells only use this mechanism in extreme stress. Y-polymerases are expressed as part of the SOS response - a coordinate response that is triggered by large-scale damage to the DNA or cell.