Adaptive Immunity - part 1
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
What are Humoral components?
Antibodies which are soluble proteins whose main
function is to communicate between innate and adaptive systems.
- Adaptive immune system signals innate immunity on
- In adaptive humoral response antibodies are needed to turn on classical
complement pathway
-Random Note: antibodies cause mast cell degranulation
Recap : What is Innate immunity and what are the 5 types
-no prior exposure to an organism is needed, there all the time
5 types:
- Physical barriers e.g. skin
- Chemical barriers e.g. pH or complement
- Inflammation
- Phagocytosis
- Natural Killer-cells (NK)
How have microbes evaded innate defences, e.g. of organism and the mechanism it uses.?
Microbes have evolved various mechanisms to evade the host's innate immune defenses. Here are some examples of organisms and the strategies they use:
Staphylococcus aureus:
Mechanism: Produces proteins that inhibit complement activation, a key part of the innate immune system.
How it works: S. aureus releases proteins like Staphylococcal complement inhibitor (SCIN), which blocks the complement cascade and prevents opsonization (marking for immune attack) and lysis of the bacteria by immune cells.
Mycobacterium tuberculosis:
Mechanism: Survives within macrophages by preventing phagosome-lysosome fusion.
How it works: M. tuberculosis blocks the maturation of the phagosome (a vesicle containing the pathogen) within macrophages, allowing it to evade degradation and survive inside immune cells.
Salmonella enterica:
Mechanism: Alters its outer membrane to resist recognition by Toll-like receptors (TLRs) on immune cells.
How it works: By modifying its lipopolysaccharide (LPS) structure, Salmonella can avoid detection by TLRs, reducing the immune system's ability to recognize and respond to the infection.
Influenza virus:
Mechanism: Uses antigenic variation to evade immune detection.
How it works: The virus frequently changes its surface proteins, hemagglutinin (HA) and neuraminidase (NA), making it difficult for the immune system to recognize and respond effectively to repeated infections.
Herpes Simplex Virus (HSV):
Mechanism: Produces proteins that inhibit the activity of natural killer (NK) cells.
How it works: HSV releases viral proteins that block NK cell activation, preventing these immune cells from identifying and destroying infected cells.
The above is from CHATGPT - double check info from MIMS Chapter 4, 11, 12, 15
What type of proteins are antibodies and how were they first detected?
-Soluble serum proteins
-First detected by electrophoresis
What is the Distribution of major human antibodies in serum protein fractions
- Most abundant = albumin
- 2nd most abundant soluble protein Gamma globulins (i.e. antibodies) = IgG (immunoglobulin G - also used as a vaccine component)(predominantly) and IgA
- Beta globulins = IgM (Bold ones are most significant)
What is the basic chain structure of Antibodies?
-Antibodies have variable amino acids for specifity and recognition
-4 polypeptide chain: 2 light chains and 2 heavy chains joined together by disulphide linkages (Disulphide bond position differs giving different t 1/2 and Vd influencing
how flexible they are)
-Y shape -Characteristic of IgG
-Fragments for antigen binding (Fab) - N terminal region that binds onto anything identified as non-self
-First crystallised (Fc region) - C terminal region has a different function - constant amino acid loops
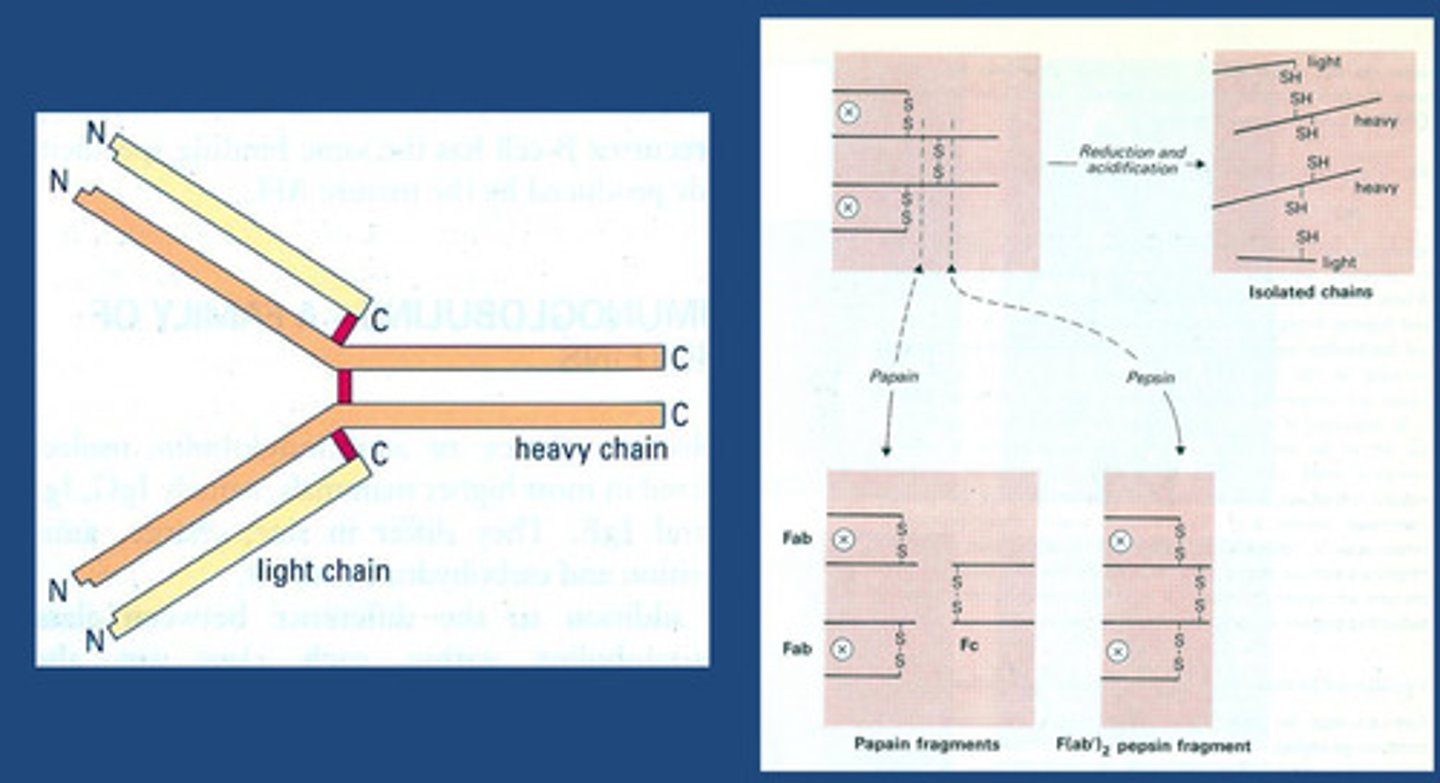
The Structure of IgG1 (a subclass of IgG) in More Detail
-Protein made up of primary amino acid that will fold on itself until it gives us a quaternary structure
-We are focused on the secondary structure:
We see loops called IgG1 molecule
--Fragments for antigen binding (Fab) - N terminal region that binds onto anything identified as non-self (i.e. antigen)
-Antibodies = variable amino acids for specifity and recognition
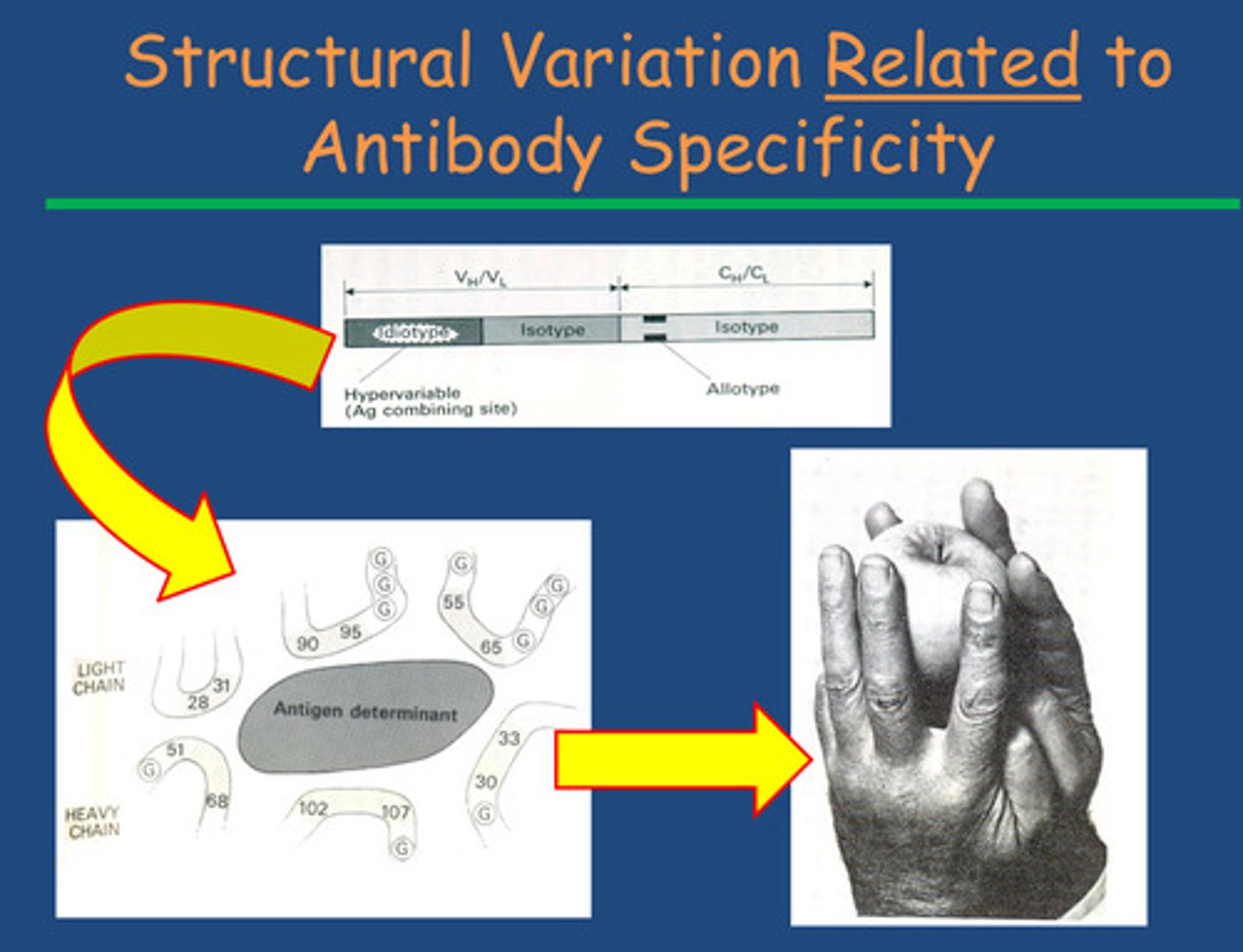
How do antibodies have specificity so you only produce the ones
that come across non-self-proteins?
- Happens by interaction of amino acids by folding of the idiotype region around the antigen = proteins from antigen interact of
antigen binding site of antibody = non-covalent bonding
- Idiotype end-region of an antibody is highly variable to allow specifity
-Isotype end of an antibody varies from species to species (in the C terminal region)
-Allotype a genetic variation in the constant regions of immunoglobulin (Ig) molecules - shows individual variation within a species' antibodies
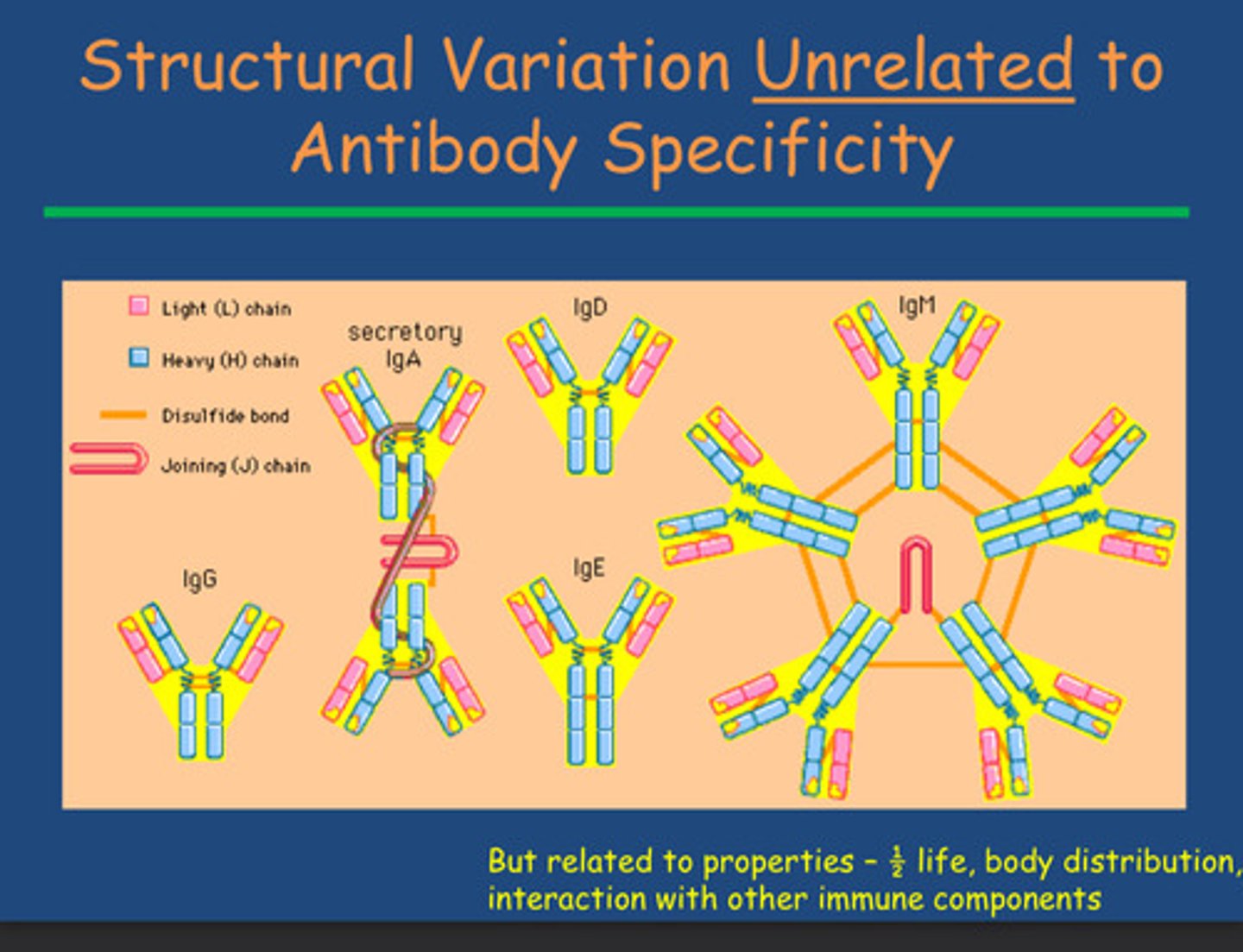
How do we get IgA, IgD and IgE?
-secretory IgA - 2 IgG together with disulphide linkages
-IgD and IgE - position of disulphide linkages
How is IgM linked to IgG and what is its function?
- IgM is a pentamer of IgG = increases number of antigen binding sites (10 in comparison to IgG's 2)
- = more likely to bind pathogens overcoming infection faster
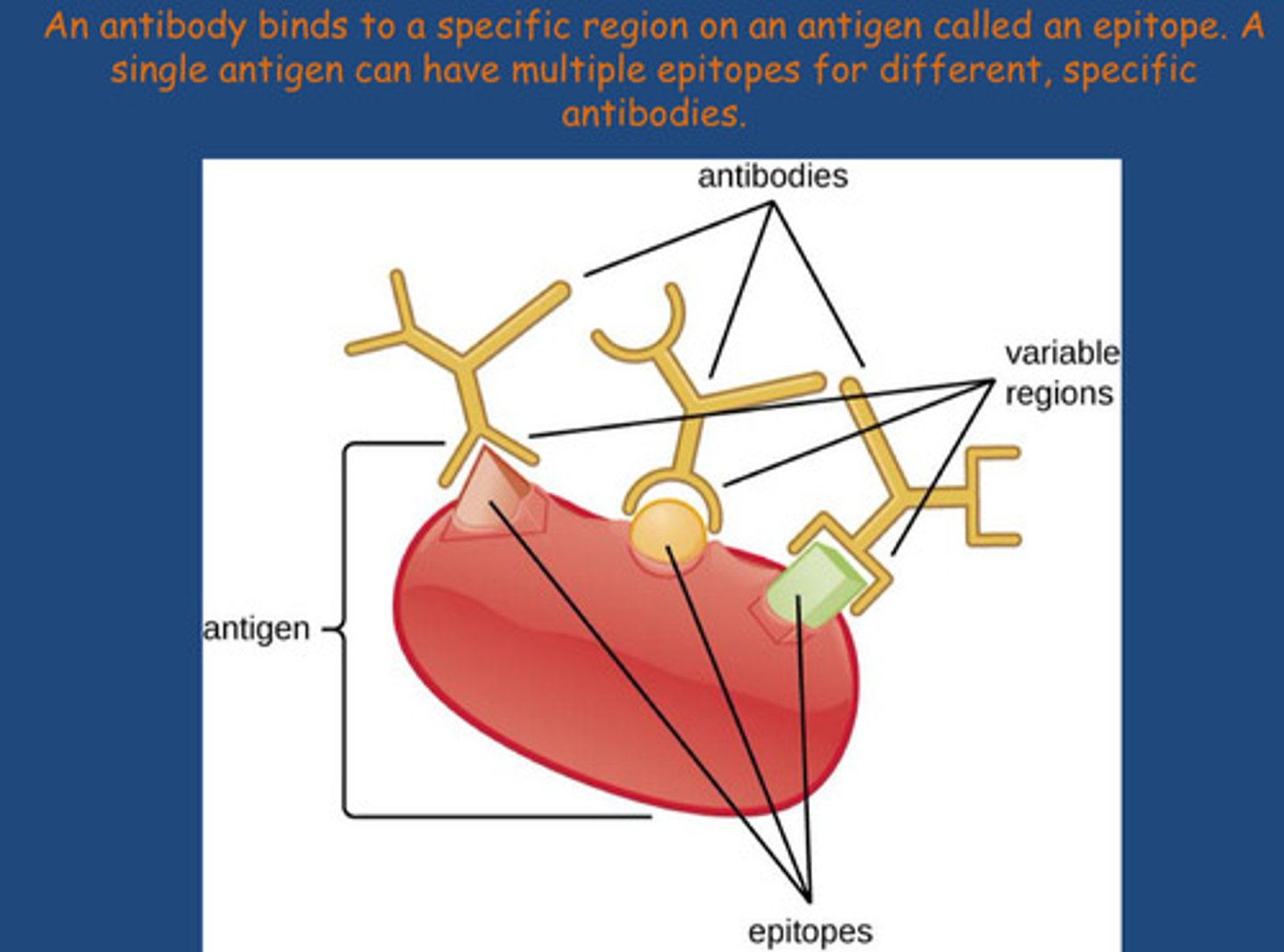
How does the body identify non-self cells ?
Antibodies recognise cells that are non self due to the difference in amino acid sequence
-Non self (antigens) is identified at the N-terminal sequence of an amino acid
-Lots of potential targets for antibodies to bind to on a specific antigen = lots of antibodies can be raised to many areas of an antigen (areas are called epitopes), in each area the amino acid sequence will be different therefore different antibodies will be produced - good to maximise the immune response from vaccines
How are antibodies produced?
- B-cells contact non-self-protein (an antigen) acting as the infectious antigen
- Progenitor haemopoietic stem cells leave bone marrow and migrate to
primary lymphoid tissues in spleen to mature into B-cells
- B-cells activate in presence of antigen to differentiate and proliferate
- Plasma cells (lot of ER = protein synthesis to secrete antibodies) and
Memory B cells (adaptive nature of immunity) - in the secondary lymphoid tissue for the next time you come into contact with a given antigen
What is the Clonal Selection theory?
- Born with B-cells
- Migrate to spleen and lymphoid tissue and wait for antigen
- Surface of B cells have B cell receptors specific to a given antigen
- One B cell will only produce an antibody with specificity for one antigen
- Antigen binding to a B-cell stimulates a specific clone to make antibody
fitting into receptor= B-cells activate in presence of antigen to differentiate and proliferate into:
- Plasma cells (lot of ER = protein synthesis to secrete antibodies) and
Memory B cells (adaptive nature of immunity) - in the secondary lymphoid tissue for the next time you come into contact with a given antigen
- Key thing: one B cell interacts with one type of antigen to give one antibody
to give surface specificity
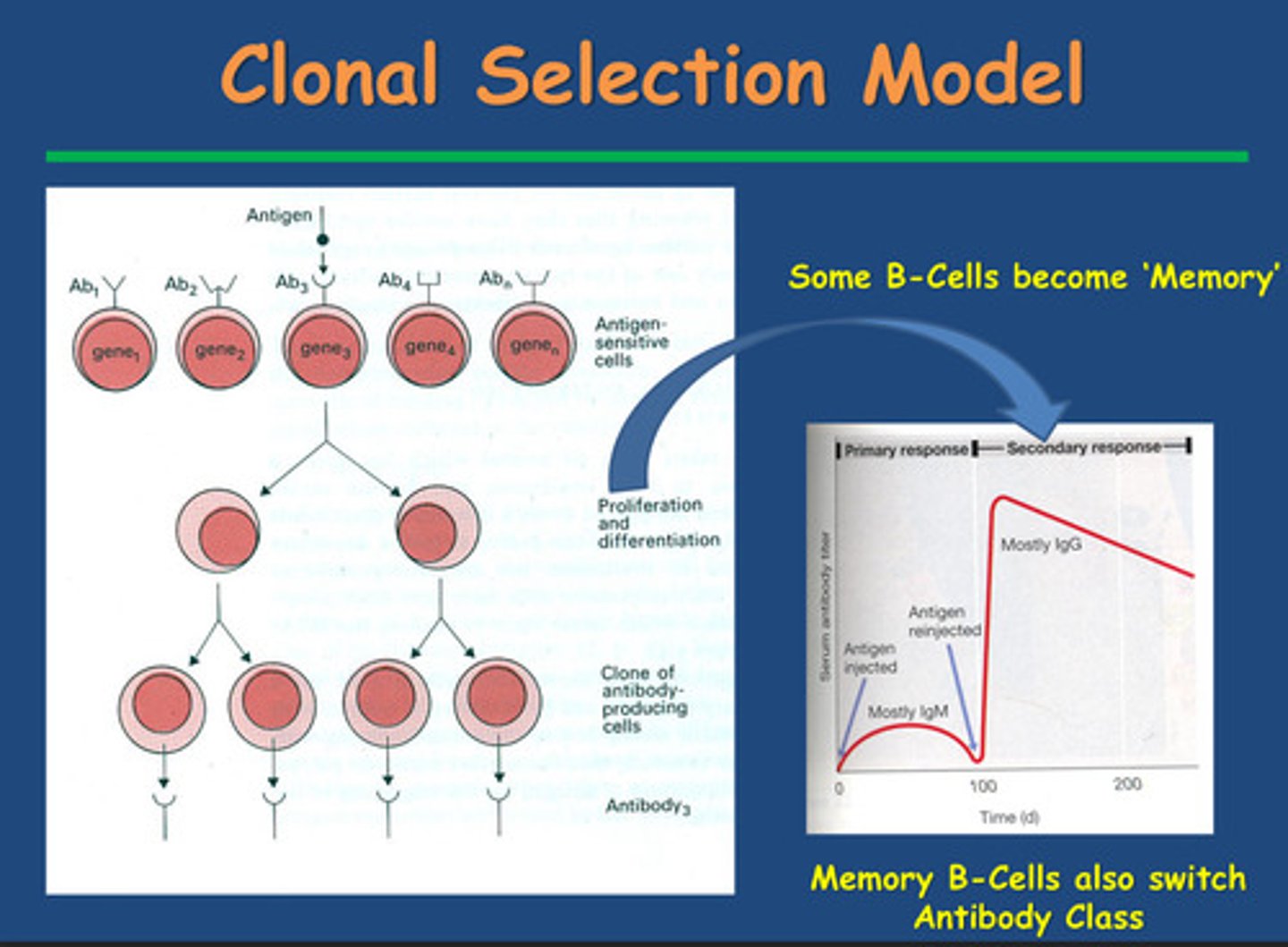
The first time you encounter an antigen, what antibody is secreted by plasma cells?
IgM
Note:
- Takes a long time to build up a large number of B-cell
clones to secrete enough antibody to be detected
- First antigen contact needs time to build up B-cells to
make antibodies
The 2nd time you encounter an antigen, what antibody is secreted by plasma cells predominantly?
IgG
-rapid antibody response due to laying down
memory cells and we see switch from IgM to IgG
What are the Characteristics of the humoral component of the adaptive immune response
-Memory - long lasting
-Specificity
-Tolerance - antibody isn't going to cross react with any other epitope o our own cells
How do we generate Antibody Diversity?
Gene Segment Recombination:
V(D)J Recombination: Antibody genes are assembled from multiple gene segments – Variable (V), Diversity (D) (in heavy chains only), and Joining (J) segments. During B cell development, these segments randomly recombine to form a unique sequence for each antibody. This recombination is catalyzed by the RAG-1 and RAG-2 enzymes.
This recombination creates a large variety of unique antigen-binding sites.
Combinatorial Diversity:
Heavy and Light Chain Pairing: Antibodies are made of two heavy and two light chains. Different combinations of heavy and light chains further increase diversity, as each unique pairing creates a different antigen-binding site
Junctional Diversity:
Addition or Deletion of Nucleotides: When the V, D, and J segments recombine, the enzyme TdT (Terminal deoxynucleotidyl transferase) adds random nucleotides at the joining regions. Additionally, nucleotides can be randomly removed. This process adds to the variability in the final antibody sequence.
Somatic Hypermutation:
After encountering an antigen, B cells undergo somatic hypermutation in their variable regions. This introduces point mutations at a high rate, particularly in the antigen-binding (hypervariable) regions. Cells producing antibodies with higher affinity are selected through a process known as affinity maturation.
Class Switch
Recombination (CSR):
Although not directly adding to the binding diversity, class switch recombination allows B cells to change the constant region of the antibody (e.g., switching from IgM to IgG), adapting the antibody's function and location in the immune response.
The above is from CHATGPT - double check info from MIMS Chapter 4, 11, 12, 15
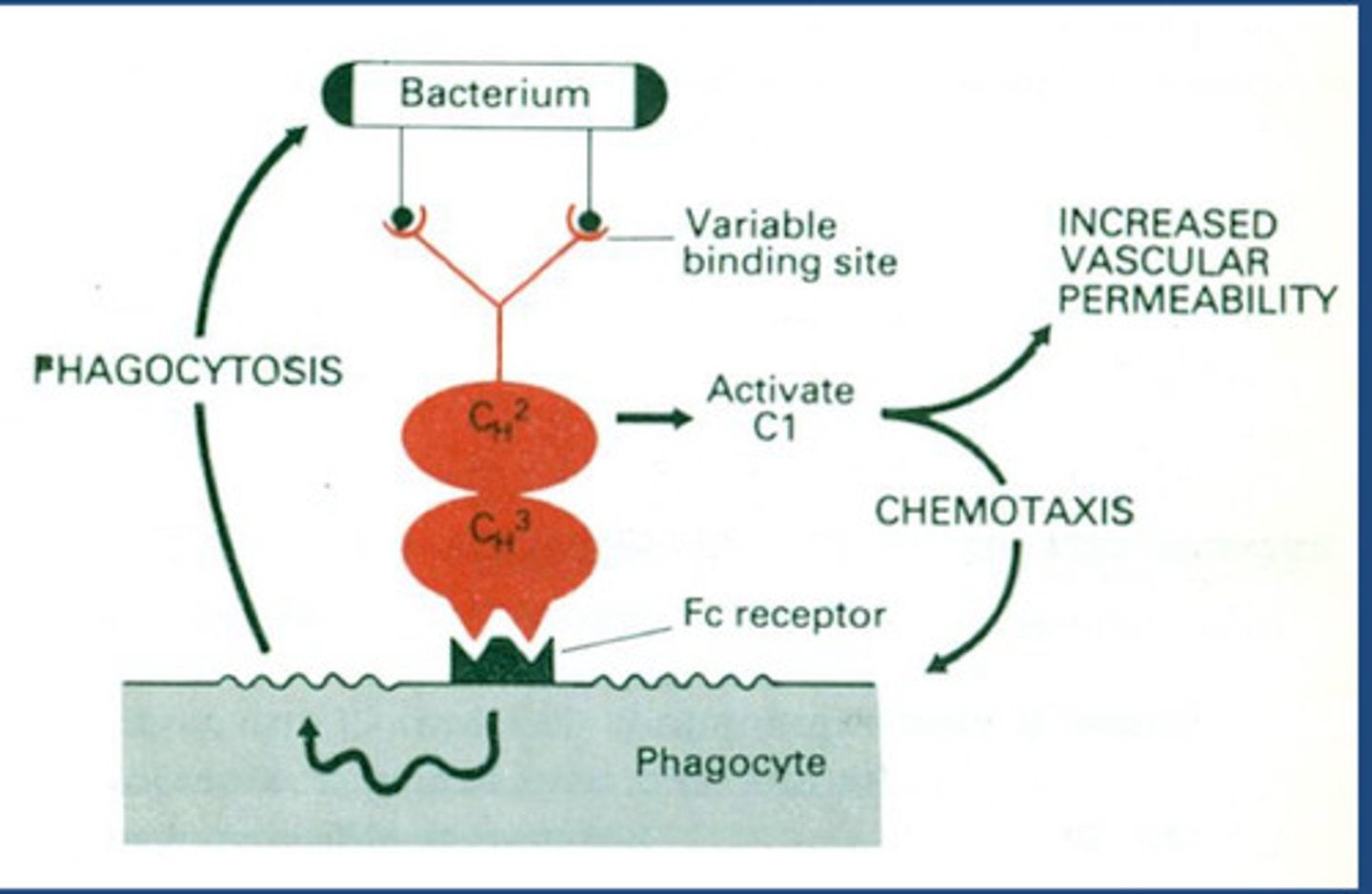
How does the antibody link to innate and adaptive immunity?
- N-terminal region has variable binding site depending on antibody type
- Bacteria have surface antigens that antibody recognises and binds =
opsonisation
- C-terminal region is constant region giving same function with an Fc
receptor
- Phagocytes also have Fc receptor
- So phagocytes detect opsonised antibodies by detecting Fc region
- Stimulates phagocytosis of bacteria = moved from adaptive to innate
- Now we have interaction between antibody-mediated activation of
complement pathway called a classical complement pathway and because it happens in constant
region all antibodies can do this, which will turn on complement= Released chemoattracted compounds - inflammatory immune response increasing
vascular permeability and phagocytosis through chemotaxis response for
polymorphic nucleocytes
Note:
- C3a activates chemotaxis
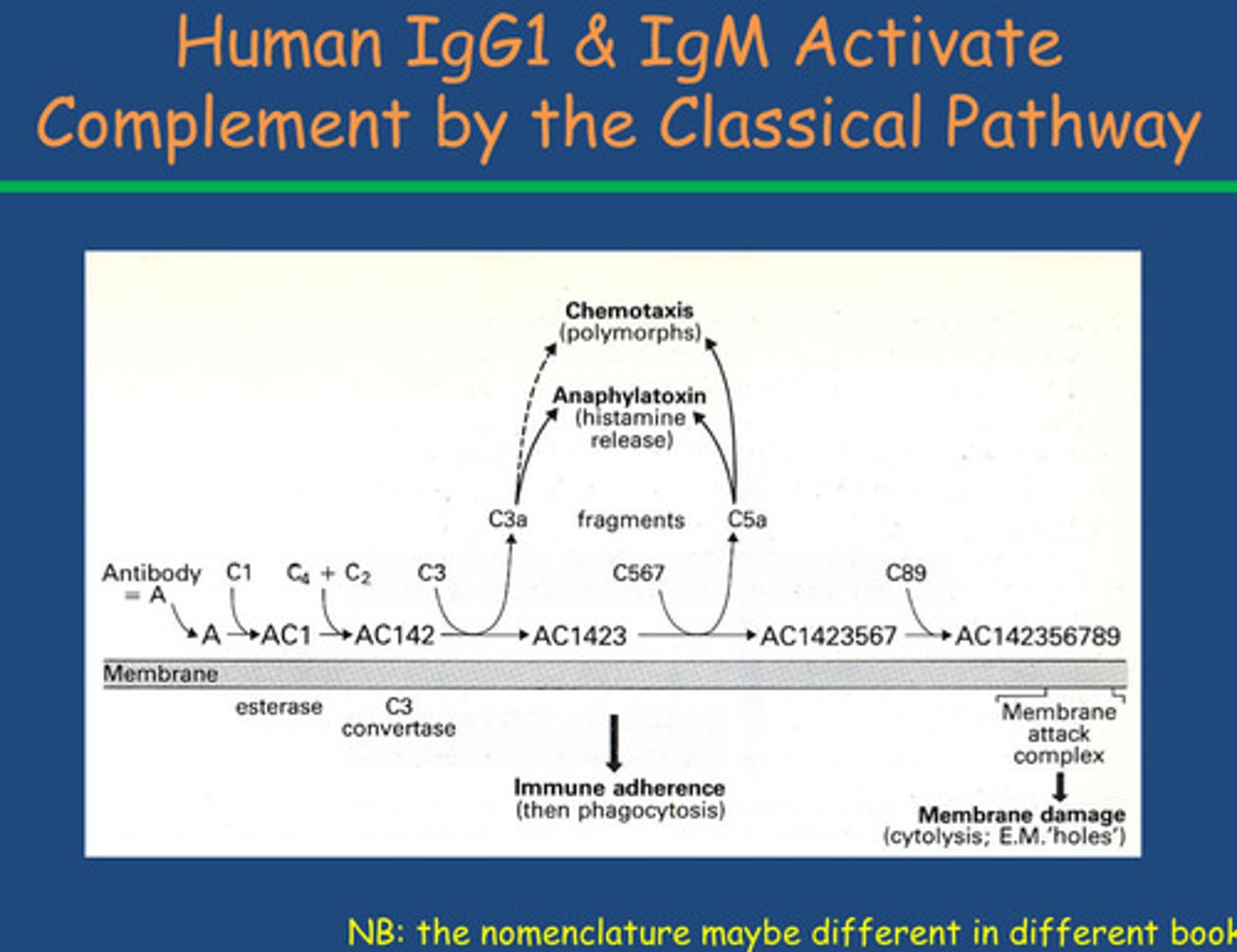
Describe and explain the Complement by the Classical Pathway
- N-terminal region has variable binding site depending on antibody type
- Bacteria have surface antigens that antibody recognises and binds =
opsonisation
- Opsonisation increases phagocytosis due to
increased adherence
- Amplify innate response by using antibodies to coat antibodies and
complements
- Need presence of bacterial interaction with C3D causing protein cascade
and C3 convertase catalysing breakdown to C3a and C3b
- Antibody must be present first- mediated by antibody
- When it binds to microbial surface get binding of C1 = antibody C1
-C4 and C2 bind to this complement protein = produces
esterase enzyme known as C1, 4 and 2 which is equivalent to C3 convertase (function above)
- C3a is chemotaxic and histamine released mast cell degranulation
-C3a will also bind to C3 convertase (this is in previous lecture)
-Gram negative organism : C5 release C5a in the Prescence of C5 convertase (chemotaxic) = mast cell degranulation also and membrane attack complex
(C5 also releases C5B in the Prescence of C5 convertase, C5b form a complex with C8 and C9 that puts a hole in the membrane of gram negative organisms - we don't know how this works)
These are different proteins but they are orthologues proteins (similar functioning proteins with genes in different species) to what we see in the alternative complement cascade
2 pathways for humoral response for complement
-Classical pathway - have to have antibody present (Step 1)
-Alternative pathway - has to have the presence of the microbial cell surface for C3b to bind on to
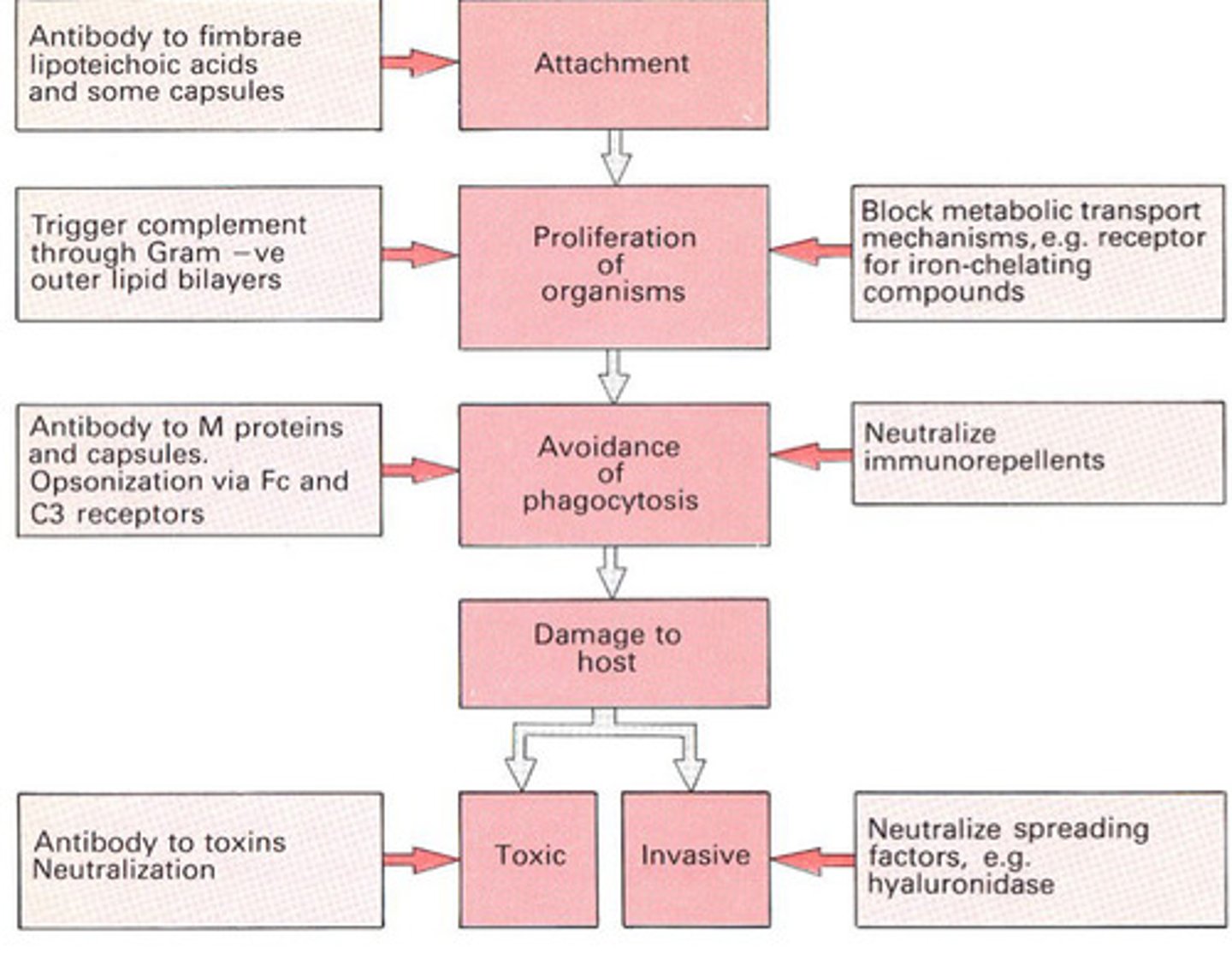
Summary - Antibody Defences Against Bacterial Infection