BIOL 3000 Chromatin and DNA Packaging
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
How many chromosome pairs are in each cell?
23
How many meters of DNA is in each cell?
~2 meters
Chromatin?
A complex of DNA and protein (histones) found in eukaryotic cells
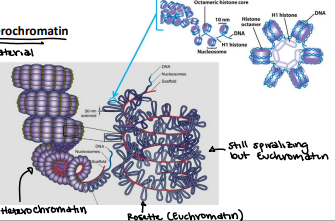
Euchromatin
Lightly packed chromatin rich in gene concentration and is most often under active transcription
Heterochromatin
Tightly packed chromatin consists mainly of genetically inactive sequences. Not a lot of genes are used
Constitutive heterochromatin
VERY gene poor; centromeres and telomeres
ALWAYS constitutive
Facultative heterochromatin
Can be gene silencing; Barr bodies
Has the ability to be both euchromatin or heterochromatin
Coding regions
Regions we are actually ablet to read
Folded Fiber Model (E.J. Dupraw)
Has whole mounts of human white blood cells
Found few or no free fiber ends and concluded each chromatid must consist of a single fiber
Extensive folding of Type B forms chromatin
Replication from each end toward the centromere
Thought to have folded upon itself, like yarn
Issues to the Folded Fiber Model
E.J. Dupraw did not give any reasons to how some chromatin went from here to there
Nucleosome Model (R.D. Kornberg, P. Oudet)
Most commonly accepted model for DNA packaging
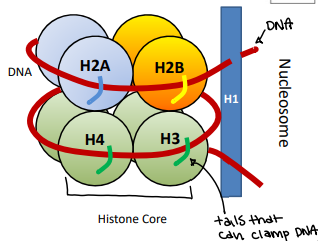
Nucleosome
The simplest packaging structure of all eukaryotic chromatin localized areas of transcription; better fit for protein biosynthesis
Has a protein core that has DNA that wraps aroundH
Histone
Protein that is heterochromatin and constitutive. There are two types: core and linker
How many histones are in a nucleosome
9
Core histones
H2A, H2B, H3, H4 (2x of each)
Consists of approximately 120 amino acids each
Highly conserved during evolution
In combination with the core particle
VERY basic in charge (approx. 25% Lysine and Arginine)
Linker histone
H1
NOT PART OF CORE but part of nucleosome
Consist of approximately 200 amino acids
Tissue specific expression and not highly conserved
Loosely associated with core particle
Only one in nucleosome
Formation of the nucleosome
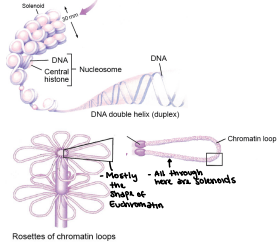
Primary packaging of the chromatin (least packaging reduces DNA length) 200 bp of DNA (146bp wrap the core; 54 bp link to the next nucleosome - linker DNA)
Reduces DNA length by ~7 times
Linear arrangement
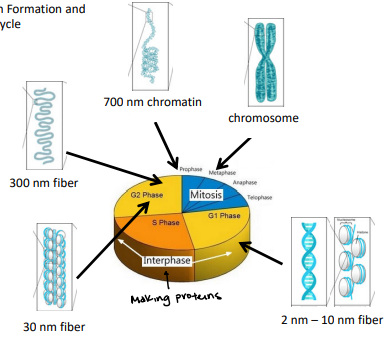
10 nm Fiber
10nm Fiber
Primary packaging of the chromatin
Supercoiling of DNA - Formation of the solenoid
H1 Histone responsible for packaging
Solenoid/30nm Fiber
Supercoiling reduces length of DNA by ~7 times
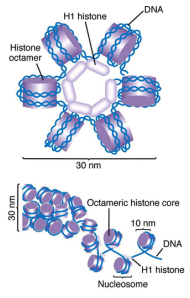
Solenoid
30nm Fiber
Helical coiling of 10nm fibers consisting of 6 nucleosomes (54 histones)
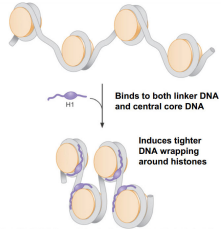
H1 Histone function
Binds to two distinct regions: linker DNA and a portion of the 146 bp core
Will compact DNA to 40x
Binds to both linker DNA and central core DNA and induces tighter DNA wrapping around histones
Supercoiling of DNA - Formation of the Zig-Zag model
The DNA backbone is not flexible enough to bend between the nucleosomes, so the straight linker DNA connects opposite nucleosomes. The zig-zag allows for greater compaction.
DNA is in the middle and does not have bends on the side
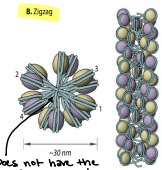
Can both solenoid and zig-zag topologies be present in chromatin fiber simultaneously?
Yes, it is experimentally shown. They are both legitimate models
Higher order coiling
300nm
“Chromatin loops”
Built around a scaffold of Topoisomerase II
Compaction level of Euchromatin (allowing us to have access to the material)
Final Condensation
700nm Fiber
Metaphase chromosome
DNA diameter is ~700nm
A spiral scaffold composed of Topoisomerase II and about 15 non-histone proteins
Compaction level of heterochromatin (cannot access DNA material)
Chromatin Formation and Cell Cycle
Karyotype
Picture of metaphase chromosomes in the nucleus of eukaryotic cells
Based off of chromosome size, centromere position, heterochromatin v.s. euchromatin