Topic 2: Inorganic Chemistry II - Transition Metal Ions in Biological Systems
1/65
Earn XP
Description and Tags
unfin
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
66 Terms
Essential Elements for Life
Include carbon, hydrogen, oxygen, nitrogen, minerals, and electrolytes like sodium, potassium, and calcium
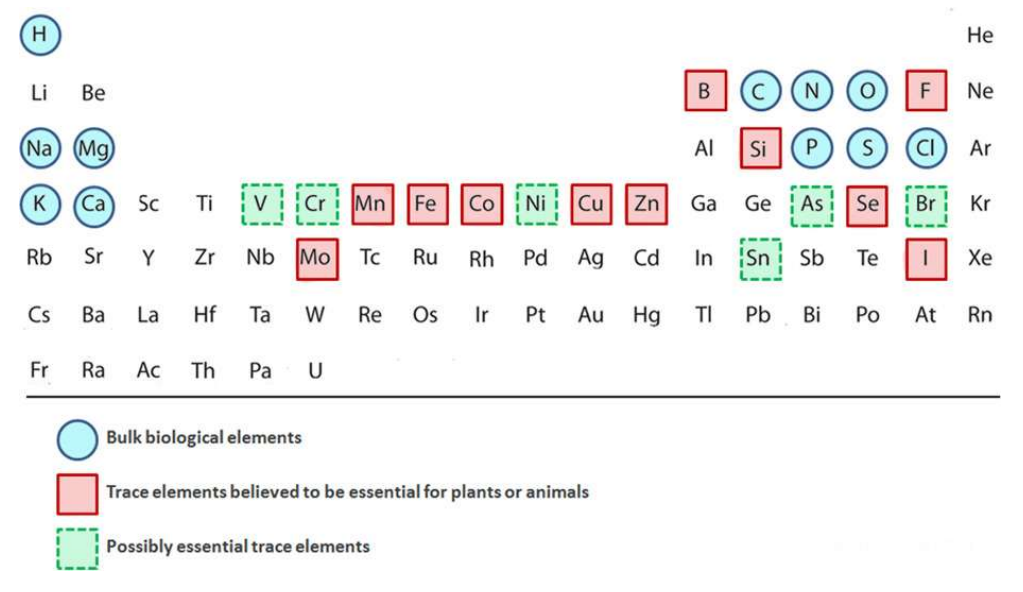
Transition Metal Ions
Constitute about 0.1% of the atoms in the human body as trace elements
Nine essential transition metals are V, Cr, Mn, Fe, Co, Ni, Cu, Zn, and Mo
Metalloproteins
Form when transition metal ions are natural constituents of proteins
Why are Transition Metal Ions Used in Biology?
They readily change oxidation states, allowing them to pick up and release electrons useful for redox chemistry
Three essential roles of metalloproteins
Transport and Storage
Enzymes (metalloenzymes)
Redox Reagents
Proteins
Polymers made of amino acids linked by peptide (amide) bonds
Protein structures
secondary (alpha helix, beta sheet), tertiary (folded shape), quaternary (dimers/tetramers)
Specific shape of a protein
designed to catalyze one specific reaction
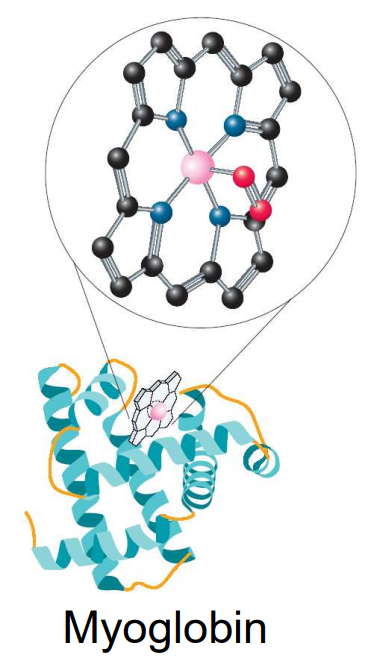
Movement and storage of oxygen
accomplished by iron-containing proteins hemoglobin (in blood) and myoglobin (in muscles)
Iron transport protein
transferrin
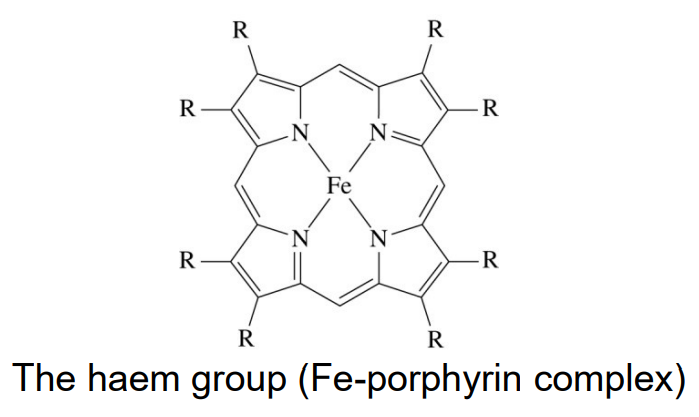
Haem group
iron-porphyrin complex acting as tetradentate, square planar ligand that chelates the iron
Hemoglobin structure
tetramer containing four haem groups, making O₂ about 70 times more soluble in blood
Deoxy-haemoglobin
contains Fe²⁺ (d⁶ High Spin), paramagnetic
Oxy-haemoglobin
iron center changes to diamagnetic upon O₂ binding, best understood as low spin Fe³⁺ (d⁵) antiferromagnetically coupled to superoxide (O₂⁻)
Carbon monoxide toxicity
CO binds more strongly to hemoglobin than O₂, displacing oxygen
CO₂ interacts with protein periphery to promote O₂ release in tissues
Cytochromes
iron-containing proteins with haem groups that facilitate electron transfer by cycling iron between Fe(II) and Fe(III)
Photosystem II
converts H₂O → O₂ + 4H⁺ + 4e⁻
contains multiple Mn atoms and one Ca atom
Manganese centers
cycle through oxidation numbers Mn(II), Mn(III), Mn(IV) to deliver four electrons
Superoxide dismutase (SOD)
enzyme that destroys reactive superoxide ion (O₂⁻)
SOD reaction
converts O₂⁻ to O₂ and peroxide (O₂²⁻ → H₂O₂)
SOD metals
uses Cu and Zn
Role of Zn in SOD
structural, holds complex in place
Role of Cu in SOD
electron shuttle cycling between Cu²⁺ and Cu⁺
Step 1 of SOD reaction
Cu²⁺–SOD + O₂⁻ → Cu⁺–SOD + O₂
Step 2 of SOD reaction
Cu⁺–SOD + O₂⁻ → Cu²⁺–SOD + O₂²⁻ (as H₂O₂)
Role of protein superstructure
protects complex and determines molecule access to metal center
Ligand fine tuning
ligands adjust the metal center to catalyze one specific reaction efficiently under mild biological conditions
Cisplatin
cancer drug cis-[PtCl₂(NH₃)₂], square planar geometry, only cis isomer is active
Cisplatin discovery
1965, Barnett Rosenberg, Michigan State University
platinum compounds prevented bacterial cell division
FDA approved 1978
Cisplatin mechanism
hydrolysis forms [Pt(NH₃)₂(OH₂)Cl]⁺, which coordinates to DNA via N7 of guanine/adenine
Cisplatin-DNA adduct
kinks DNA helix, blocks DNA repair, causes cell death
Technetium (Tc)
no stable isotopes, all radioactive
Technetium-99m
metastable isomer used in medical imaging (e.g., heart blood flow)
Technetium coordination chemistry
Tc ions combined with ligands to form injectable complexes for targeted imaging
Chelation therapy
treats toxic metal buildup using chelating agents to form excretable complexes
Chelating agent mechanism
binds selectively with toxic metals to form complexes removable by kidneys
Chelating ligands
usually polydentate with high formation constants (β), stabilized by chelate effect
Ligand selection
guided by hard/soft acid-base principles
Chelation challenges
chelates may remove essential metals, requiring supplements
EDTA⁴⁻
common but non-selective chelating agent
DMSA (meso-2,3-dimercaptosuccinic acid)
removes soft metals like Hg²⁺ and Pb²⁺ using sulfur donors
Deferasirox
reduces acute iron levels (e.g., thalassemia)
Fe³⁺ prefers hard donors (O, N) present in deferasirox
Oxidation state calculation
metal charge = complex charge − sum of ligand charges (e.g., [Re(CO)₅Cl] → Re⁺¹)
IUPAC definition of transition element
element with an incomplete d subshell or forming cations with incomplete d subshells
Zinc exclusion
Zn not a transition element because Zn and Zn²⁺ have full d¹⁰ configurations
d-electron count
formula = (Group number − Oxidation state)
Coordination sphere
octahedral (6-coordinate) or square planar (4-coordinate)
Bidentate ligands
form two bonds to metal (e.g., ethylenediamine, oxalate)
iodide is not bidentate
Octahedral crystal field splitting
d orbitals split into t₂g (dxy, dxz, dyz) and eg (dx²−y², dz²)
High Spin (HS) vs Low Spin (LS)
determined by Δ₀ vs pairing energy (P)
If Δ₀ < P
complex is high spin
If Δ₀ > P
complex is low spin
Electron counts capable of HS/LS
d⁴, d⁵, d⁶, d⁷
Spectrochemical series
order of ligand field strength: halides < oxygen donors < nitrogen donors < carbon donors
Magnetism
diamagnetic if all paired, paramagnetic if unpaired electrons present
Magnetic moment formula
μ = √(n(n+2)) μ_B
Jahn-Teller distortion
occurs in d⁹ configurations (e.g., Cu²⁺) due to partial filling of e_g orbitals
Cause of Jahn-Teller distortion
degenerate e_g orbitals split into two energy levels when distorted
Result of Jahn-Teller distortion
energetically favorable stabilization by lowering energy of partially filled orbital