LO3: Development of DNA Sequencing Technologies
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
46 Terms
Molecular Biology
a field of biology that studies the composition, structure, and interactions of cellular molecules that carry out the biological processes essential for the cell’s function and maintenance
Central Dogma
Illustrates the flow of genetic information from DNA to RNA to Protein
DNA Replication
process by which a double-stranded DNA molecule is copied to produce two identical DNA molecules
Steps of DNA Replication
Unwind, Prime, Elongate
DNA Polymerase
The enzyme responsible for constructing new DNA strands during replication or DNA repair.
Can only elongate DNA where there is a free 3’ -OH group that it can act on.
DNA Helicase
The enzyme that unwinds (unzip) the DNA molecule near the replication fork
Single-Stranded DNA-Binding Proteins (SSBs)
Are proteins that bind to and stabilize the single-stranded regions of DNA that result from the action of unwinding protein
Topoisomerase
It facilitates DNA replication by reducing molecular tension caused by supercoiling upstream from the replication fork (e.g. DNA gyrase)
Primase
Is a polymerase that initiates replication by synthesizing short segment of RNA as source of the 3’-OH end that DNA polymerase can use.
DNA Ligase
It seals nicks or breaks in the sugar-phosphate backbone generated during replication
Polymerase Chain Reaction
a laboratory technique generating tens of billions of copies of a particular DNA fragment (the sequence of interest, DNA of interest, or target DNA) from a DNA extract (DNA template)
Requirements for PCR
DNA Extract
Polymerase (Thermus aquaticus)
Primers
4 dNTPs
Buffer Solution
Thermal Cycler
Basic Principles of PCR
Denature, Anneal, Extend
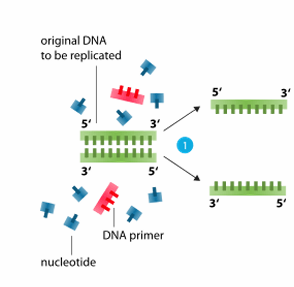
Denature
separation of two strands of DNA
carried out at a temperature of 94°C
double-stranded DNA is denatured into single-stranded DNA (H-bonds denature)
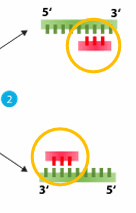
Anneal
primer hybridization temperature
carried out at a temperature generally between and 40 and 70°C
uses Primers
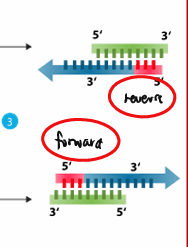
Primers (Anneal)
short, single-stranded sequences complementary to regions that flank the DNA to be amplified
Extends
carried out at a temperature of 72°C
synthesis of the complementary strand
Polymerase binds to the primed single-stranded DNAs and catalyze replication using the dNTPS present in the reaction mixture
Agarose Gel Electrophoresis
a procedure that reveals stained DNA via ultraviolet transillumination (280-320 nm)

Analysis and Techniques Based on PCR
Microsatellites
Single nucleotide polymorphism (SNPs)
Amplification of fragment length polymorphism (AFLP)
Restriction fragment length polymorphism (RFLP)
Mitochondrial DNA polymorphism (mtDNA)
Variations of PCR
Reverse transcriptase PCR (RT-PCR)
Quantitative PCR in real time (quantitative real-time PCR)
Semi-quantitative or competitive PCR
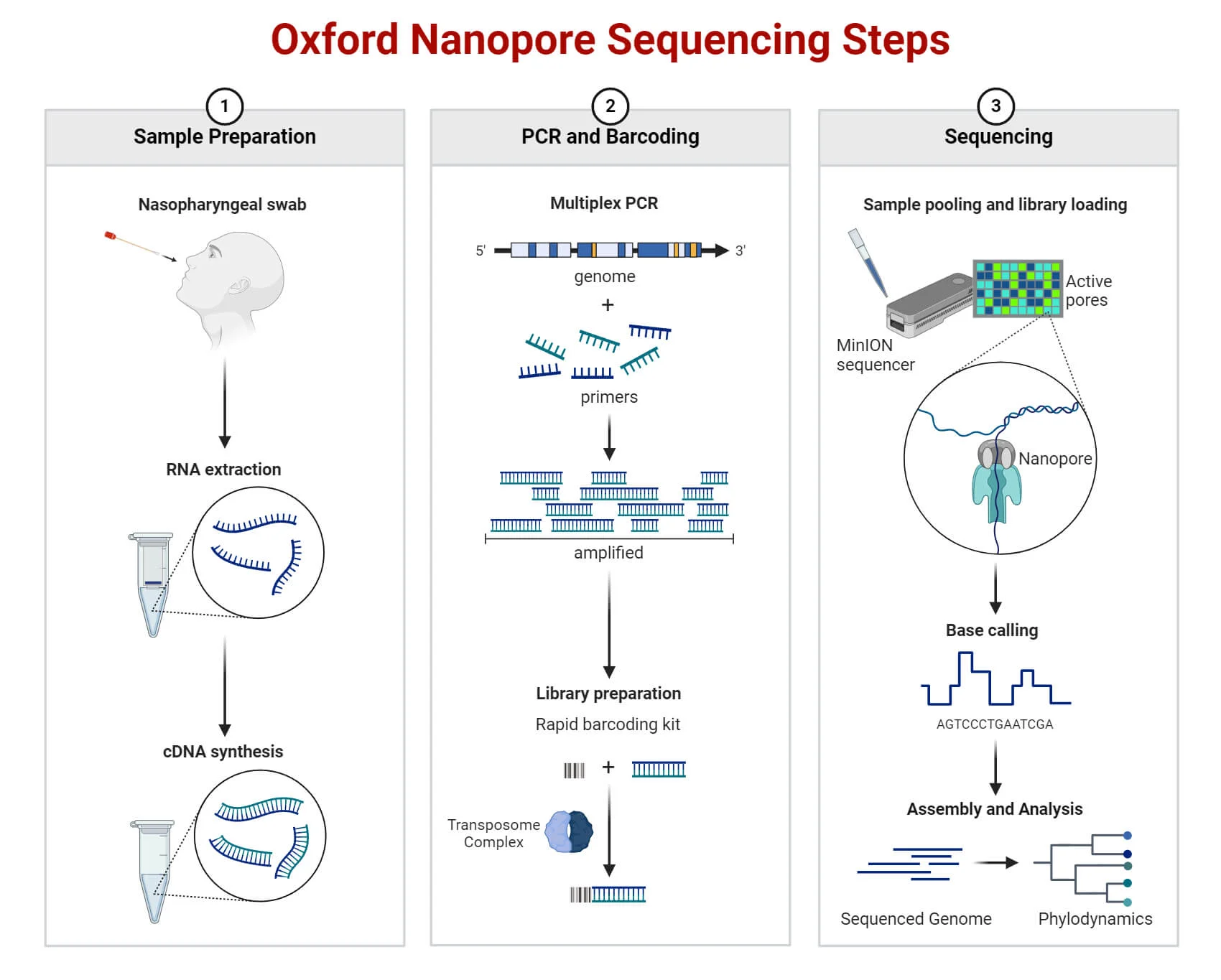
DNA Sequencing Technologies
First Generation, 2nd Generation, 3rd Generation
1st Gen Sequencing
Maxam-Gilbert Sequencing
Sanger Sequencing
2nd Gen Sequencing
454 Pyrosequencing
Illumina Sequencing
SOLiD Sequencing
Ion Torrent Sequencing
3rd Gen Sequencing
PacBio RS System
Nanopore Sequencing Technology
1st Gen | Maxam-Gilbert Sequencing
sequencing by chemical degradation requiring chemical modifications of the DNA and further cleavage and electrophoresis
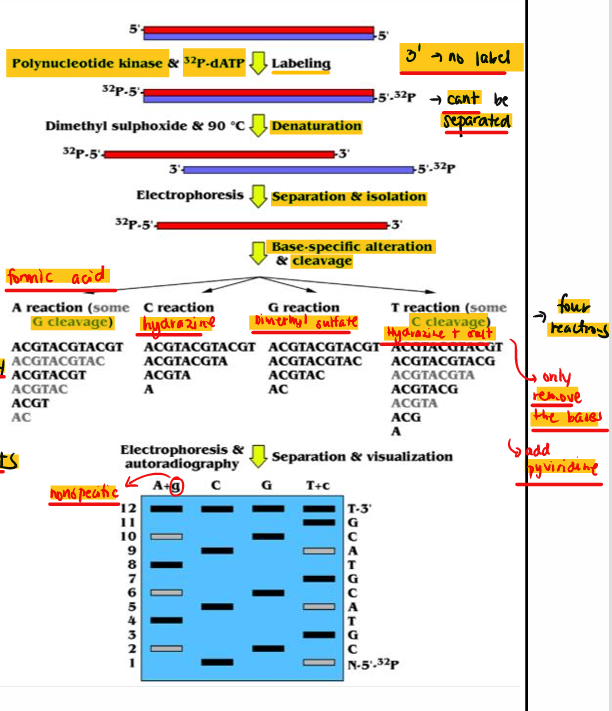
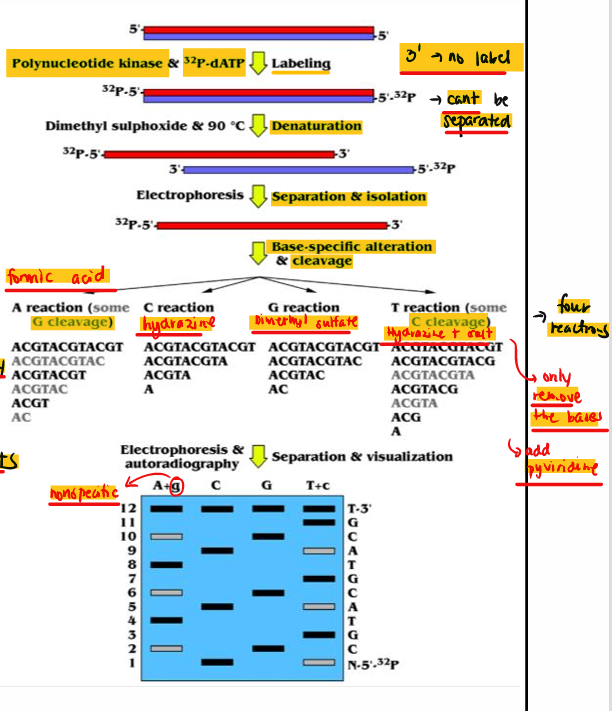
[STEPS] Maxam-Gilbert
Radioactive labeling of the 5’-P ends of dsDNA with 32P-dATP
Denature with DMSO at 90°C then electrophoresis
Modification of the nitrogenous bases
Chemical cleavage of the ssDNA at the 5’-P side
Electrophoresis and autoradiography
Infer DNA sequence
[DISADVANTAGES] Maxam-Gilbert
Uses hazardous chemicals
Technically challenging
Difficult to scale-up
1st Gen | Sanger Sequencing
a nucleic-acid sequencing approach by enzymatic synthesis, aka chain termination method
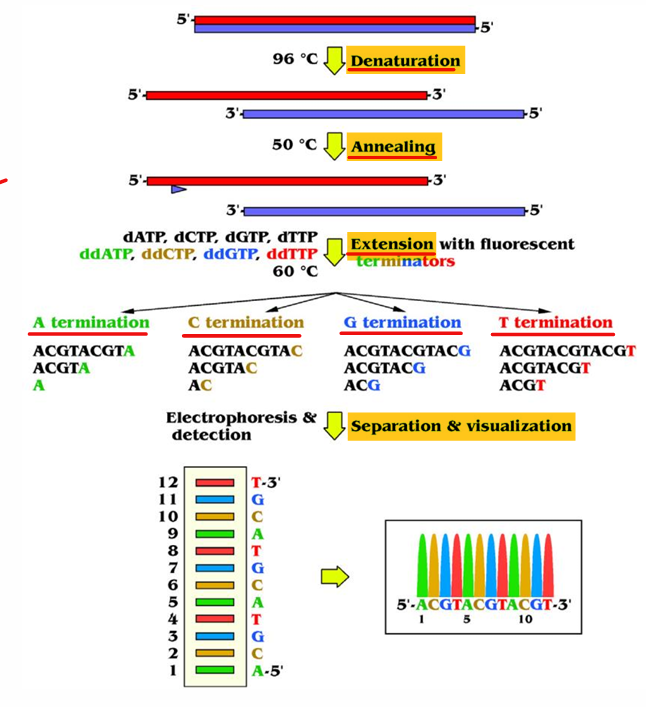
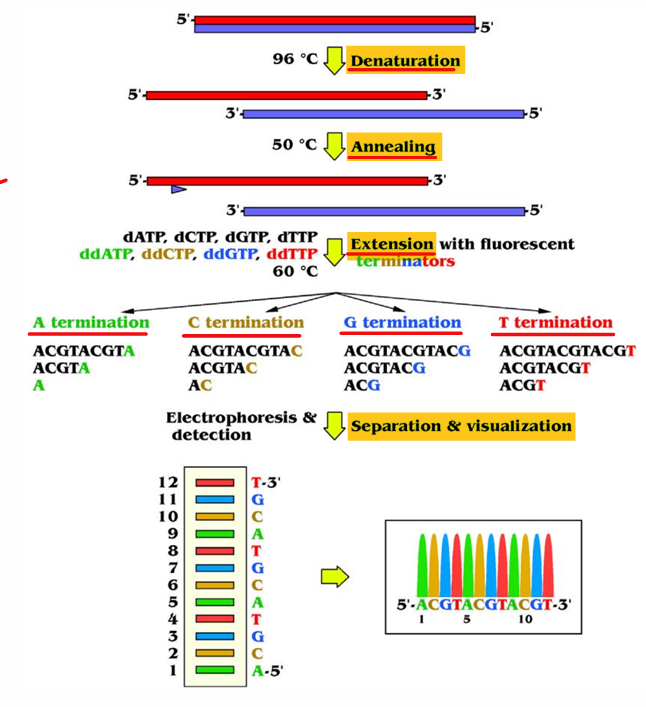
[STEPS] Sanger Sequencing
Denature the dsDNA
Anneal with primers
Extend with dNTPS and ddNTPs
Segregation via gel or capillary electrophoresis
Interpret virtual electropherogram or electrofluorogram
[DISADVANTAGES] Sanger Sequencing
Expensive (equipment, reagents, workforce)
Time consuming
Not suitable for genomic sequencing
2nd Gen | Roche 454 Pyrosequencing
emulsion-PCR technology generating the longest reads (from 200 to 1000b)
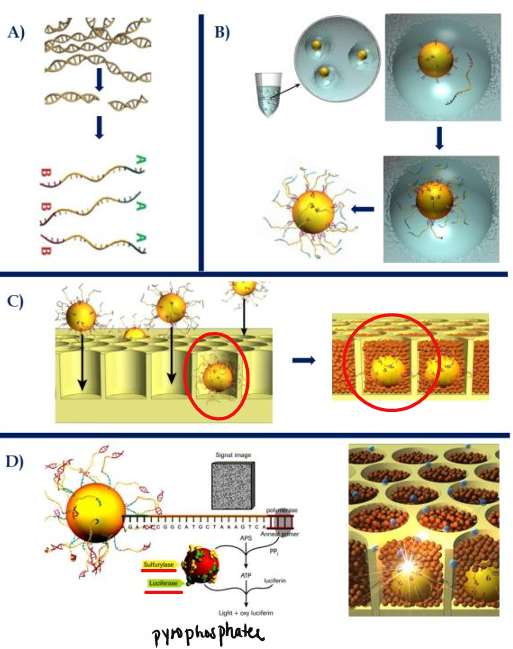
[STEPS] Roche 454 Pyrosequencing
Library construction = nebulization, adapter ligation, biotin-tagged DNA attaches to streptavidin in beads
Emulsion PCR
DNA-enriched beads to picotiter wells + smaller beads containing enzymes (ATP sulfurylase, luciferase, and apyrase)
Single nucleotide is flown, signal detection (base calling), wash/degrade remaining bases
Sequencing = image/signal processing
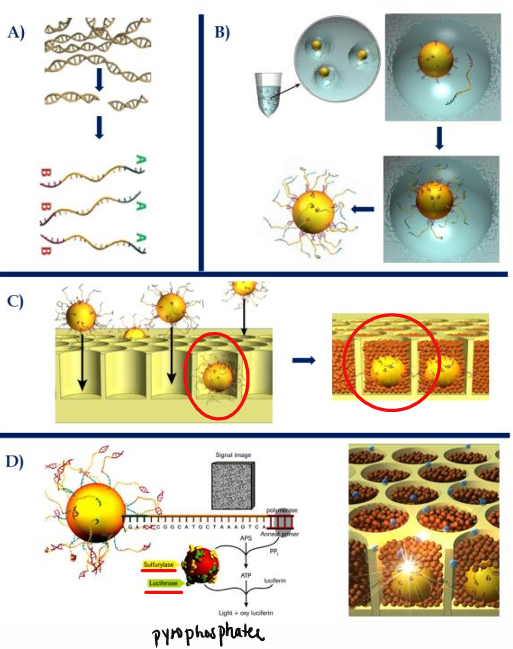
[DISADVANTAGES] Roche 454 Pyrosequencing
Homopolymeric regions —> InDel errors
Challenging sample prep
2nd Gen | Illumina Sequencing
Reversible-terminator sequencers generating shorter reads (35-150b) but larger genome coverages
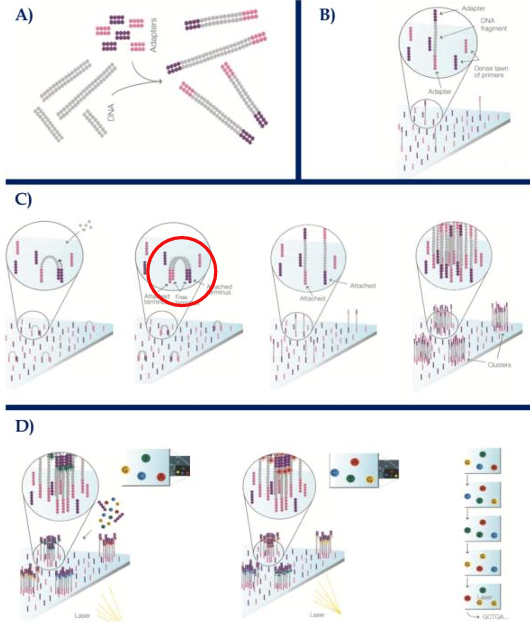
[STEPS] Illumina Sequencing
Library construction = fragmentation, ligation of “Y” adapters, immobilization
Cluster generation = PCR or bridge amplification
Sequencing = reversible-terminator sequencing using fluorescent nucleotides
Data Analysis = assemble the reads into contigs
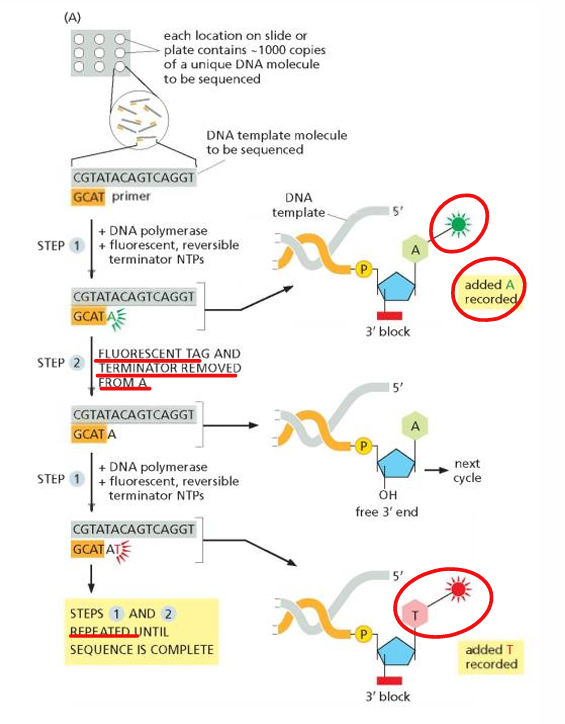
[DISADVANTAGES] Illumina Sequencing
Substitution errors
Signal decay (lower accuracy at ends)
2nd Gen | SOLiD Sequencing
Sequencing by Oligonucleotide Ligation and Detection (SOLiD) can also generate a high coverage, yet with short reds (25-75 bases)
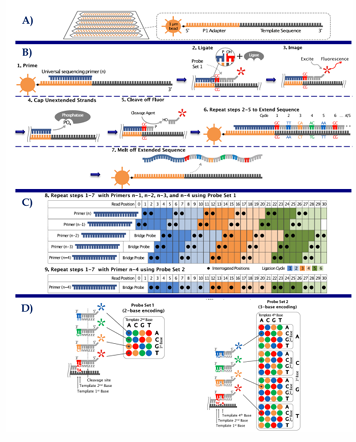
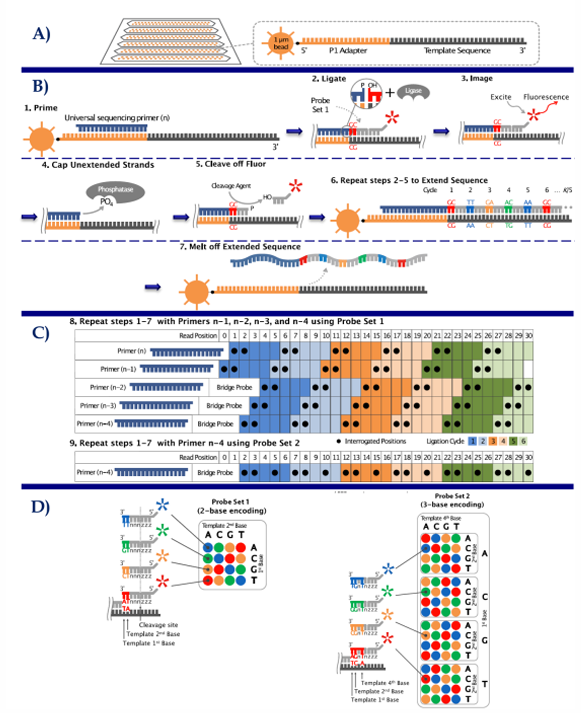
[STEPS] SOLiD Sequencing
Library Construction —> emPCR —> DNA immobilized in glass surface
Sequencing = five universal sequencing primers (with 2 interrogative bases) —> ligate —> fluorescent probes = repeated in n-1, n-2, n-3, & n-4
Data Analysis = assemble the reads
[DISADVANTAGES] SOLiD Sequencing
Highly complex
Challenging sample prep
2nd Gen | Ion Torrent Sequencing
based on the detection of the protons generated on the polymerization reactions of nucleic acids
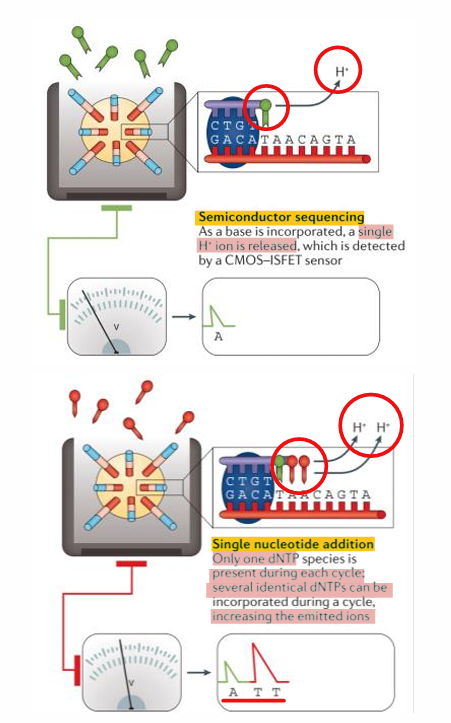
[STEPS] Ion Torrent Sequencing
Library construction —> emPCR —> DNA immobilized in picowells
Sequencing = one - by - one dNTP-flush extension detection the proton released
Data Analysis = assemble the reads
[DISADVANTAGES] Ion Torrent Sequencing
Still relies on clonally amplified template
Errors in homopolymeric regions
3rd Gen | PacBio SMRT Sequencing
Single-molecule real-time (SMRT) sequencing using special loop adapters and zero mode wavelength design (ZMW)
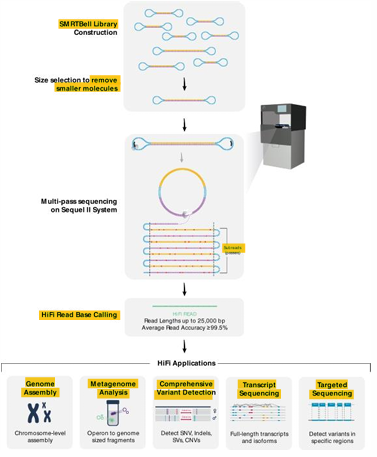
[STEPS] PacBio SMRT Sequencing
Library construction = follows strand displacement amplification (rolling circle)
Sequencing = fluorescently labeled base (P) —> light emission
Data Analysis = assemble the reads
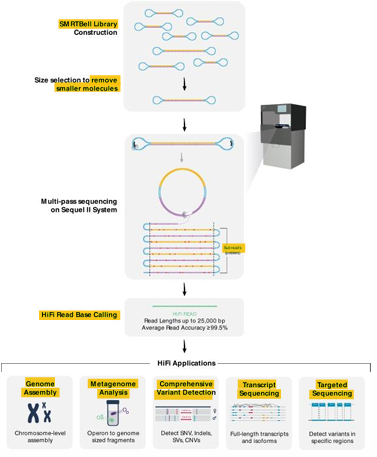
[DISADVANTAGES] PacBio SMRT Sequencing
Very high error rates
Requires high molecular weight template
Costly
3rd Gen | Nanopore Sequencing
based on the detection of the nitrogenous bases as they move through pores embedded in a lipid bilayer on microwells