Nucleic acids
1/64
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
65 Terms
Nucleotide
A pentose sugar bonded to a phosphate and a nitrogenous base.
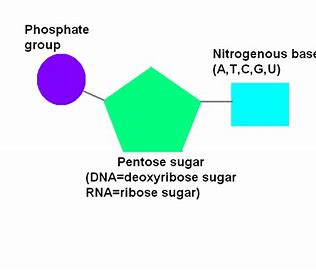
Nucleoside
Pentose sugar joined to a nitrogenous base. E.g. adenosine (part of ATP) is made of ribose bonded to adenine.
DNA (full name and components)
Deoxyribonucleic acid. Consists of deoxyribose sugar and a base out of Adenine, Guanine, Cytosine and THYMINE.
RNA (full name and components)
Ribonucleic acid
Made of sugar ribose, a phosphate group and base one of Adenine, Guanine, Cytosine or URACIL.
ATP (full name and components)
Adenosine triphosphate. Made of ribose sugar, base adenine and three phosphate groups
ADP (full name and components)
Adenosine diphosphate. Made of ribose sugar, adenine base and 2 phosphate groups.
ADP to ATP (5)
Phosphorylation
Endergonic reaction; uses energy released during cellular respiration.
ADP + Phosphate (Pi) → ATP
Condensation reaction
catalysed by ATP SYNTHETASE.
Endergonic vs exergonic reactions
Endergonic- Uses energy
Exergonic- releases energy
Condensation reaction
Bonds formed through the removal of water
Hydrolysis reaction
Bonds broken through the addition of water.
ATP to ADP (4)
ATP can be hydrolysed
to form ADP + Pi.
Exergonic: Releases 30.6kJ/mol of energy.
Reaction catalysed by enzyme ATPase.
ATP as universal energy currency.
It provides energy to all biochemical reactions in all cells in all species by the hydrolysis of the phosphate-phosphate bond
Significance of ATP (6)
nature of reaction
enzyme(s) involved
size of energy released
transportation of molecule
type of energy produced
uses of energy produced
energy released in a single hydrolysis reaction
catalysed by a single enzyme (ATPase)
Energy released in small, usable quantities of 30.6kJ/mol (unlike glucose)
Small and soluble, so easily transported into cells.
Convert energy into a single form, the same energy can be used for all biochemical reactions in all cells of all living organisms
Energy needed for processes like active transport, protein synthesis, cell division and muscle contraction
Where is ATP synthesised?
Mitochondria and chloroplasts
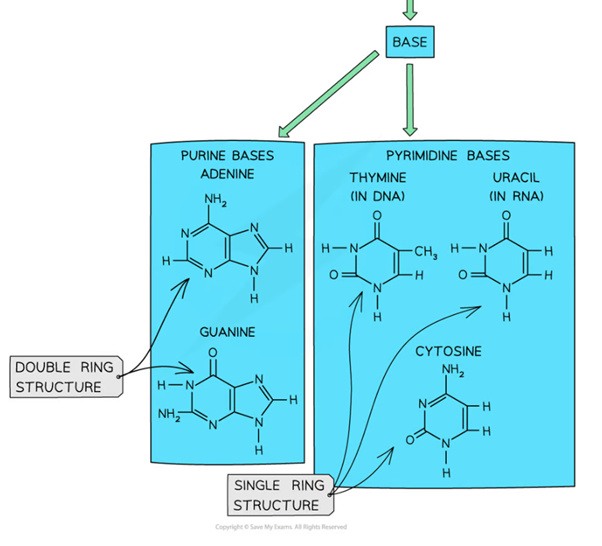
Two catagories of base, differences
Purines- have a double ring structure (Guanine and Adenine)
Pyrimidines- have a single ring structure (Thymine, Cytosine, Uracil)
Proportion of bases in DNA
Adenine: Thymine - 1:1
Guanine: Cytosine - 1:1
Purines: Pyrimidines - 1:1
Proportion of bases in RNA
RNA is single stranded so proportions of bases is random.
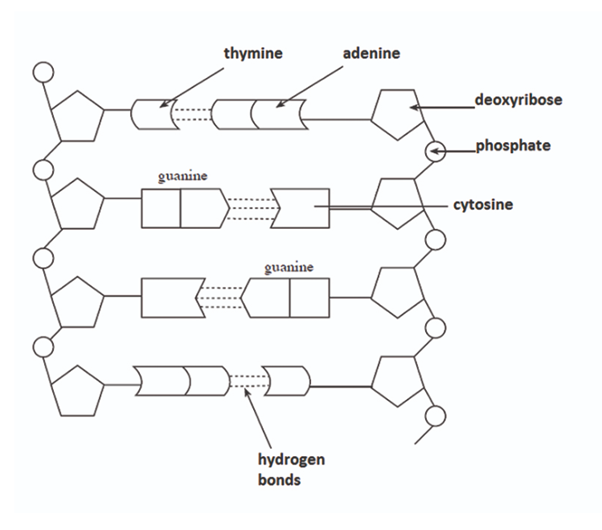
Bonding between complementary bases
Adenine and thymine are linked with 2 hydrogen bonds.
Guanine and Cytosine are linked with 3 hydrogen bonds.
Functions of DNA
Protein synthesis: as base sequence coding for amino acid sequences
DNA replication: prior to cell division to each daughter cell get equal DNA
Structure of DNA (two strands and their linkage) (4)
DNA is composed of two comlementary polynucleotide strands twisted into a double helix.
Nuclotides are joined through condensation reactions; there is phosphodiester bonds linking the phosphate group of one nucleotide and the pentose sugar of the next.
There are alternating sugar-phosphate molecules that form two anti-parallel (one from 3’ to 5’, another from 5’ to 3’) sugar-phosphate backbones of DNA.
Hydrogen bonds between complementary bases hold the two strands together.
Three types of RNA
Ribosomal RNA (rRNA)
Messenger RNA (mRNA)
Transfer RNA (tRNA)
Ribosomal RNA (3)
Made in nucleolus
Is a component of Ribosomes (as well as protein)
The single polynucleotide is highly folded to form a globular structure.
Messenger RNA where and role (3)
In the nucleus.
It is made using complementary RNA base pairs to the template DNA strand.
mRNA carries the code from DNA out from the nuclear pore into the cytoplasm where it attatches to ribosomes.
Codon
Three bases on mRNA that code for an amino acid
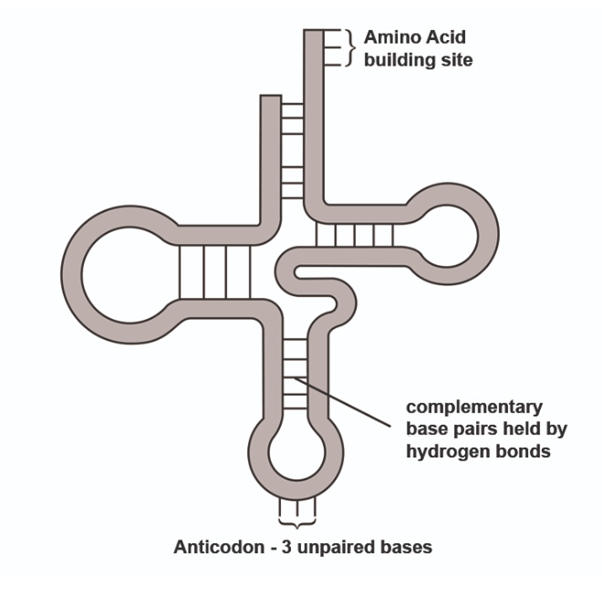
Transfer RNA
RNA twised into a clover-leaf shape, held in place by hydrogen bonds between complementary bases.
Its function is to carry amino acids to the ribosomes. The amino acids is determines by the anticodon on the ‘middle leaf’ of the clover shape. These anticodons are complementary the codons on mRNA and ensure the amino acid sequence is in the right order for the primary protein structure.
Steps for extracting DNA from cells (4)
Crush and blend the sample of cells in detergent, salt and water to release DNA. The detergent is used to dissolve the membrane of the cells to release DNA. The salt is used to form a precipitate out of the DNA.
Filter the cell debris and collect the extract into a test tube.
Pour ice-cold cold alcohol down the side of the tube containing the extract.The DNA precipitates at the junction of the extract and alcohol.
DNA can be stained red using acetic orcein.
Risk assessment for extracting DNA
Alcohol ~ Skin irritant ~ wear safety goggles
What are the three possible mechanisms for DNA replication?
Conservative replication
Dispersive replication
Semi-conservative replication
Describe the Dispersive DNA replication possibility
DNA molecule would be copied and spliced together, making each new DNA molecule a mix of the original and new DNA.
Conservative replication possibility
DNA molecule would be copied from the original, leaving the original DNA molecule as it was and having a new copy.
Semi-conservative method of DNA replication
The two polynucleotide chains in the original DNA molecule would part, and new nucleotides attach to each end of the chains, leadoing to each new molecule having one original chain and one new one.
How did Meselson and Stahl investigate DNA replication in a lab?
what means did they use to obtain DNA? Why?
How did they distinguish between original and new strands of DNA?
Meselson and Stahl used E.coli bacteria as a source of DNA. This bacteria is easily grown in a culture and replicate their DNA every 20 mins under optimal conditions.
‘New’ and ‘original’ nucleotides were distinguished by the isotope of nitrogen in nitrogenous bases. N-15 is heavier in mass than N-14, so these isotopes can be separated by mass in centrifugation.
What happened when Meselson and Stahl grew E.coli in culture of only N-15
After many generations, all of the bases contained N-15, which is heavier N. So, when centrifuged, the band of DNA was near the bottom of the tube.
What happened when a sample of E.coli that had been cultured in N-15, gets transferred to a new culture medium of N-14 and allowed to replicate once, when centrifuged?
When centrifuged, the DNA would be found at an intermediate position between heavy N-15 at the bottom of the tube, and N-14 light at the top of the tube.
How did Meselson and Stahl’s experiment disprove the conservative method.
After one division in N-14 culture after being completely in N-15 culture, there was a mix of heavy and light, so DNA replication could not be the conservative method, otherwise there would be the original band at the bottom, and new light band of DNA at the top.
How did Meselson and Stahl disprove the dispersive method of DNA replication?
The E.coli bacteria was left to replicate, forming generation 2. In this centrifuge tube, there were two bands of DNA of equal widths at the intermediate position and at the light position.
This dissproves the dispersive method of replication, as it would have to have more N14 than N15 and the centrifuged DNA would have to be between the intermediate and light position.
How did Meselson and Stahl prove the semi-conservative method of DNA replication?
The E.coli formed in the second generation showed the ‘hybrid’ (one strand of each N) molecules had parted and used as a template for the new light nucleotides to attach to, leading to the band in the ‘light’ position.
What would the third generation of centrifuged DNA show in Meselson and Stahl’s experiment?
A larger width band at the light position and a smaller width band at the intermediate position. This is because the ratio of light to intermediate is 3:1.
Role of DNA helicase in DNA replication
what is the specific name of where DNA helicase works? Where does it begin working?
How is this different for eukaryotes and prokaryotes
Unwind and unzip the double helix DNA strand by break the hydrogen bonds holding the two polynucleotide strands together (unzipping).
The area where DNA helicase works is called the ‘replication fork’. DNA helicase begins ‘unzipping’ at a place called the origin. There are typically multiple origins in the DNA of Eukaryotes, while prokaryotes only have one origin.
Role of DNA polymerase in DNA replication
Which way does DNA polymerase build in, and so which direction does it move in along the template strand?
To join new nucleotides to their complementary bases by catalysing the formation of phosphodiester bonds between deoxyribose and phosphate groups. The original polynucleotide chains act as a template for the aligning of new nucleotides.
DNA polymerase builds the new strand in the 5’ to 3’ direction, so it moves along the template strand in the 3’ to 5’ direction to make the antiparallel strands.
Primase in DNA replication
An enzyme that places RNA primers onto the DNA strand to indicate to DNA polymerase where to start building the new strand.
Leading vs Lagging strand in DNA replication
When DNA polymerase builds the new strand in the 5’ to 3’ direction, on one original strand, DNA polymerase will be moving in the same direction as DNA helicase in unzipping in, whereas on the other anti-parallel strand, DNA polymerase is moving in the opposite direction to DNA helicase.
This means that primers keep having to be placed on one strand for DNA polymerase to replicate the unzipped region. This strand is called the ‘lagging’ strand, and the other original strand is called the leading strand.
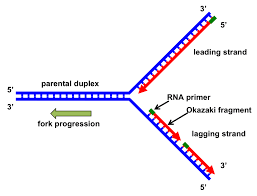
How is the lagging strand completed.
When Okazaki fragments are formed from the placement of primers along the lagging strand, the primers have to be replaced with DNA bases as the primers themselves were made of RNA.
DNA ligase then has to ‘glue’ together the okazaki fragments to remove any gaps between them.
Products of DNA replication (4)
Two
identical
semi-conservative
double helix DNA molecules
Gene
nature of genes in eukaryotes vs prokaryotes
A section of DNA that codes for a polypeptide.
In eukaryotes, genes contain introns and exons. Exons are the coding parts, and introns are the non-coding parts.
In prokaryotes, their genes are continuous with no introns.
First step of protein synthesis- unwinding
Part of transcription
DNA helicase unwinds and unzips the two DNA strands by breaking the hydrogen bonds between the two polynucleotide strands, exposing the bases. One strand acts as the template strand for the formation of mRNA.
Protein synthesis: formation of pre-mRNA
Part of transcription
RNA nucleotides align to their complementary bases and are joined together by RNA polymerase forming the ribose phosphate backbone of mRNA.
This is the pre-mRNA that is formed and RNA polymerase leaves the DNA when the stop codon is reached.
Protein- synthesis: Post-transcription modification
Involved the removal of introns that are left in the nucleus, and the splicing together of exons.
The functional mRNA strand now leaves the nucleus through the nuclear pore.
One gene one enzyme hypothesis
It was though in 1945 that genes only coded for enzymes which catalysed the production of other proteins.
How was the ‘one gene one enzyme’ hypothesis dissproved?
In 1950s studies of haemoglobin showed that genes also coded for proteins other than enzymes. Therefore the hypothesis was modified to become the ‘one gene one polypeptide hypothesis
What is the ‘one gene one polypeptide hypothesis’
and why is it thought to be too simple?
Each gene is transcribed and translated to form a single polypetide.
It is too simplistic as exons can be spliced in different orders forming different types of mRNA from one type of pre-mRNA. The different code results in different proteins. This eliminated the one gene one polypeptide hypothesis.
Codon
every three bases on mRNA
Anticodon
Three bases on tRNA that determine which amino acid is carried by tRNA.
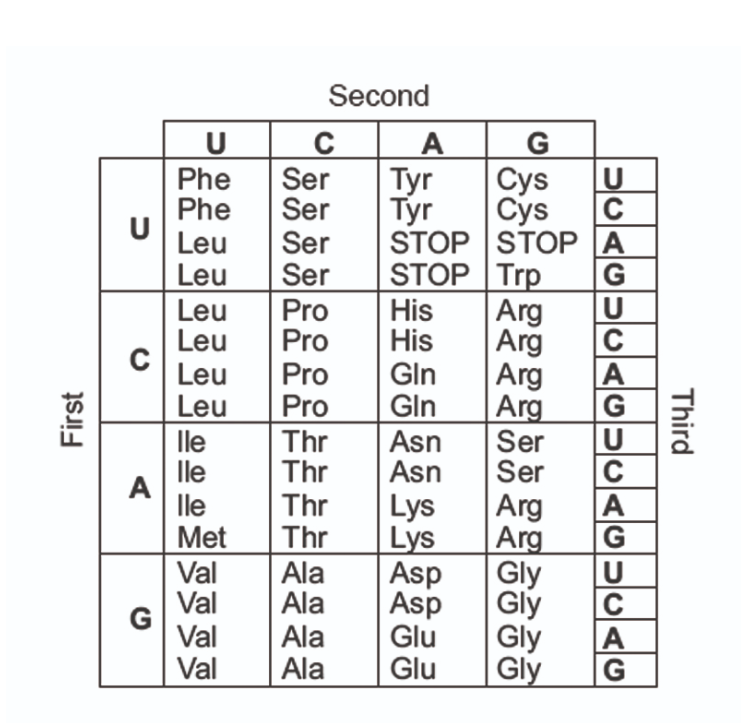
Genetic code
How can genetic code be read and interpreted?
A triplet code on DNA coding for one amino acid.
The code can be read using a genetic dictionary which is univeral for all living things.
The code is degenerate, so some amino acids have more than one code. This means that mutations may not have an affect on the amino acid.
How many codons can a ribosome accommodate?
2 codons
Protein synthesis: translation and role of tRNA
An amino acid is attached to tRNA’s amino acid binding site using energy from ATP. It is determined by the anti-codon on tRNA which binds to a complementary codon on mRNA, by forming hydrogen bonds.
When two tRNA molecules occcupy the ribosome, the amino acids form a peptide bond between them. Then the first tRNA is released from the amino acid and returns to the cytoplasm to be reactivated.
This process continues adding amino acids onto the polypeptide chain until a stop codon is reached.
What happens to the polypeptide chain after leaving the ribosome?
It enters the golgi body for proteins modifcation.
Here, the polypeptide chain can be folded to make a protein, and may hv non protein (prosthetic) groups added like carbohydrates, lipids or phosphates.
The genetic code is (6)
a
linear
triplet
non-overlapping
degenerate
unambiguous
universal code
for the production of polypeptides
linear genetic code
read in a line
triplet genetic code
how many possible combinations of triplet bases are there?
three bases on DNA code for one amino acid
43 combinations of codes= 64
non-overlapping genetic code
codons are read without overlap
degenerate genetic code
Some amino acids have more that one triplet code
unmabiguous genetic code
clear code
universal genetic code
same code in all living things
Eukaryotic genes difference from prokaryotic genes
Eukaryotic have discontinuous genes with coding exons and non-coding introns
Prokaryotic genes are usally continuous and lack non-coding sequences