immu2011: complement system
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
68 Terms
complement components
a collection of circulating and cell membrane proteins, important for defence against microbes and in antibody mediated tissue injury
why its called “complement”
ability of proteins to assist or “complement” the antimicrobial activity of antibodies
may be activated in absence of antibodies
or by antibodies attached to microbes
activtion of complement
involves sequential proteolytic cleavage of complement proteins
consequences of complement activation
generation of effector molecules that participate in eliminating microbes
regulation of complement activation
controlled by molecules on host cells to prevent harm
diseases related to complement
arise due to defects in complement function
effector molecules
are generated in large numbers during complement activation
the cascade of protein activation amplifies greatly
covalent attachment
activated complement proteins become attaches to cell surfaces (covalently) where the activation occurs → ensure activation is limited to correct sites
control of complement
tightly regulated by molecules present in normal host cells, preventing uncontrolled + harmful complement activation
pathways of activation
classical
alternative
lectin
C3 protein
most abundant complement protein found in blood
C3 convertase
formed by Cub, crucial in activating downstream pathways
C3a function
attracts leukocytes and promotes inflammatory processes
C3b function
opsonisation, aiding phagocytosis, and activation of downstream pathways
C5 cleavage by C3b
forms
C5a for inflammation
C3b for membrane attack complex
C3 tick over
continuous low rate cleavage of C3 to generate C3b
complement initiation by microbes in absense of antibodies
alternative
lectin
complement activation by antibodies
classical pathway
result of all complement pathways
all 3 pathways have same results, just different methods of reaching them
result = rid math open (make pore inside to cause osmotic lysis of pathogen)
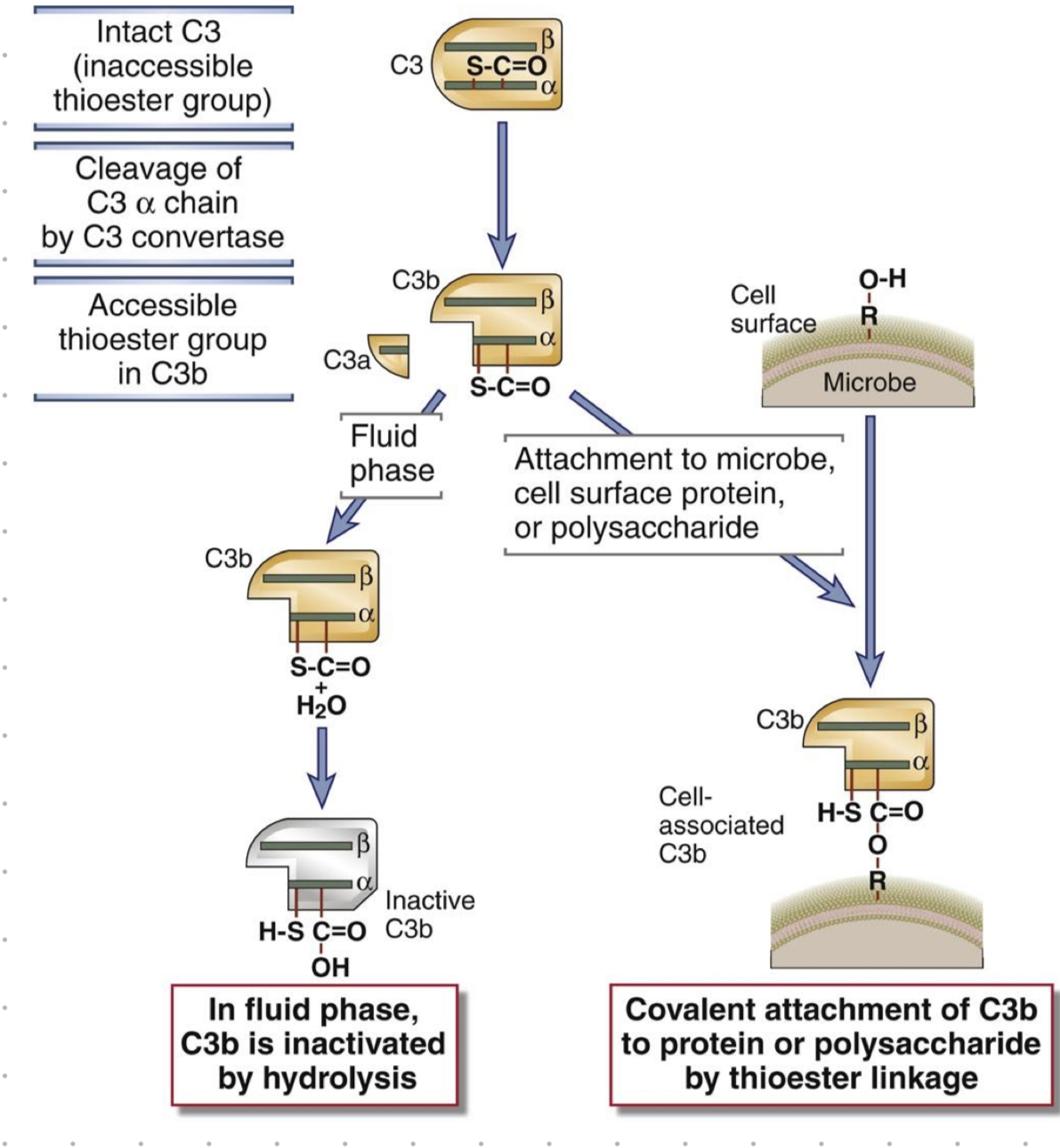
thioester group
reactive site on C3b for covalent attachment to microbes
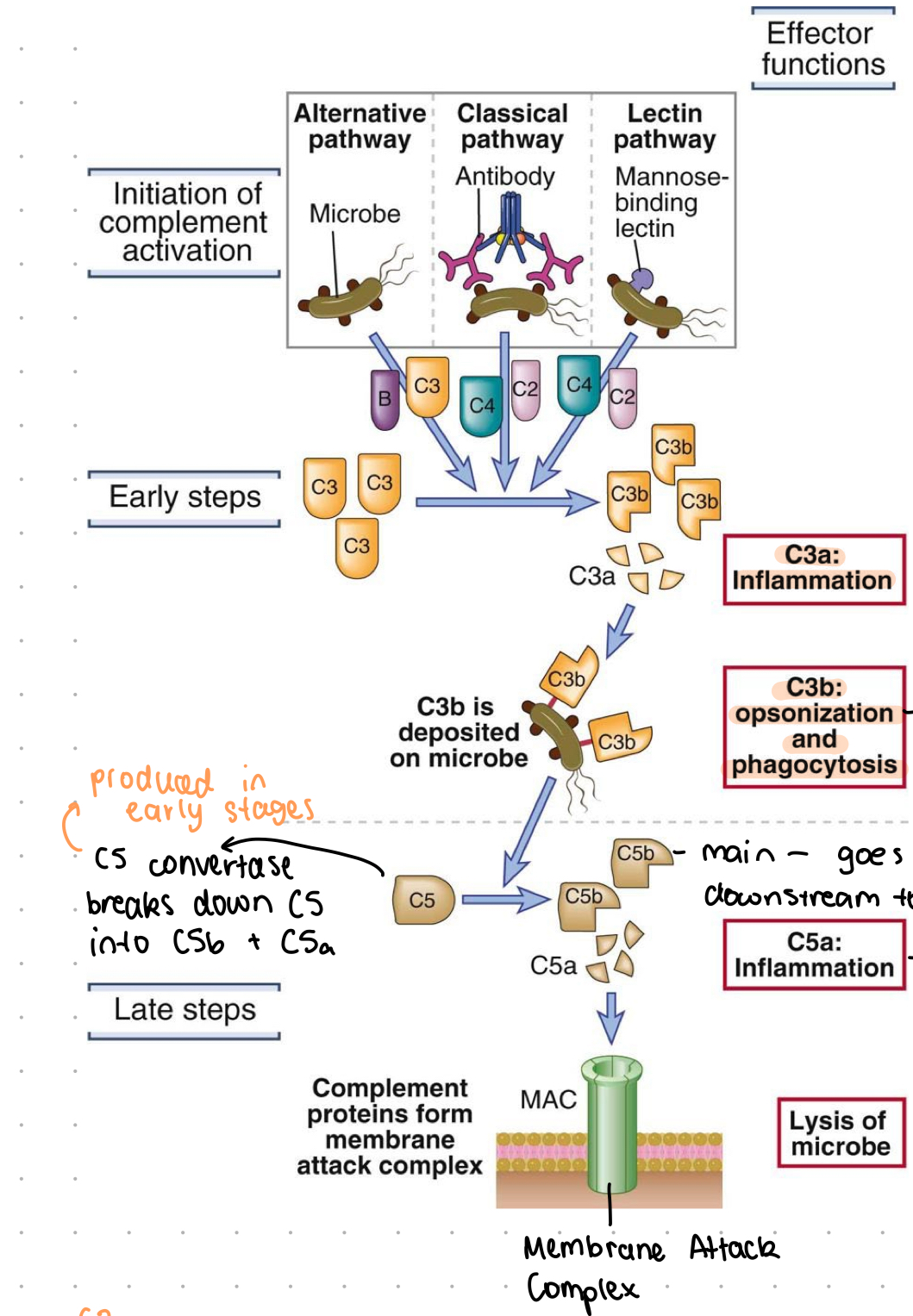
complement steps (basic)
initiation by: microbe, antibody, mannose-binding lectin
cleavage of C3 into C3b and C3a
C3b deposited on microbe, C3a promotes inflammation
C3b binds to microbe surface where it: A) inhibits it, or other receptors recognise C3b on microbe and cause phagocytosis
C5 is cleaved by C5 convertase into C5a and C5b
C5b goes downstream to form membrane attack complex, C5a promotes inflammation
lysis of microbe
inactivation of C3b
occurs if no microbe is present, prevents further activation of downstream processes
pathogen absence
leads to C3b degradation and inactivity over time
membrane attack complex
MAC
formed by C9 polymerization at C5b-8 site
embeds into lipid bilayer, forms a pore
C3 origin
always present in blood
produced by liver and enters blood at whole component
slowly degrades in blood (when no microbes) into C3b and C3-alpha
free floating C3b
present in blood, binds to microbes (alt. pathway) and complement system is activatates
if no microbes, it eventually becomes inactive
C3-alpha
result of C3 degradation in blood when no microbes present
inactive
C3 cleavage
cleavage of C3-alpha cain (by C3 convertase)
thioester group in C3b becomes accessible
in blood (when no microbes)
C3b is inactivated by hydrolysis
microbes present?
attach to microbe, cell surface protein or polysaccharide by covalent bonding
factor B
protein broken down by factor D to form Bb fragment
Bb fragment
generated by breakdown of factor B, part of C3b-Bb complex (alt. pathway)
alternate pathway - facts
used for microbes in blood
complements enter, factor B interacts w/ C3b and forms complex
factor D breaks down factor B into subunits
C3b-Bb - creates C5 convertase
cleavage of C5 and initiation of late steps of complement
C3b-Bb complex (of alt.)
enzymatically breaks down C3, acts as an alternative pathway C3 convertase
C3b-Bb-C3b complex
functions as a C5 convertase, breaks down complement protein C5 (in alt. pathway)
C5 convertase
complex that breaks down C5, initiation the late steps of complement activation
classical complement pathway
activated by antibody-antigen complexes, begins with binding of C1 to antibody-antigen complex
C1 complex
composed of C1q and tetramer of 2 C1r and 2 C1s molecules (serine proteases), initiates complement cascade
C1q = recognition component
C1r and C1s = serine proteases
Fc portion
region of antibodies that C1 complex binds to to initiate complement pathways
note - only antibodies bound to antigens, not free circulating antibodies, will initiate the pathway
antibodies which trigger complement
IgM
IgG1 and IgG3 (subclasses of IgG)
C4 (classical)
cleaved by activated C1s, generating C4b and C4a
note - second most abundant CP in blood
antibody-antigen complexes which initiate classical pathway may be…
soluble
fixed on cell surfaces
deposited on extracellular matrices
C2 (classical)
complexes with C4b, cleaved by C1s to form C2b and C2a
C4b-2A complex
classical pathway convertase cleaving C3 into C3b and C3a
C4b (classical)
contains an internal thioester bind, similar to C3b, which covalently attaches to cell surface/antigen complex
attachment of C4b ensures that classical pathway activation proceeds on a cell surface or immune complex
opsonisation
attachment of multiple C3b molecules to microbes
C4b-2a-C3b complex
functions as C5 convertase in classical pathway
classical pathway steps
binding of C1 to antibodies of antigen-antibody complex
binding of C4 to Ig-associated C1q (of C1)
cleavage of C4 by C1s enzyme into C4a and C4b
covalent attachment of C4b to antigenic surface and antibodies
C2 binds with C4b, cleaved by C1s to form C2b and C2a
C2a forms complex w/ C4b on cell surface (C4b-2a)
C4b-2a = C3 convertase, cleaves C3 into C3b and C3a
C3b opsonisation + binding to microbe
C4b2a bind to C3b → C4b-2a-C3b complex = C5 convertase, cleaves C5
lectin pathway
activated by mannose-binding lectin (MBL), recognises terminal mannose residues on microbial glycoproteins and glycolipids
has hexameric structure (similar to C1q)
antibodies required
MASP1 and MASP2
mannose associated serin proteases
similar functions to C1r and C1s
MBL binding to microbes initiates downstream proteolytic steps identical to classical pathway
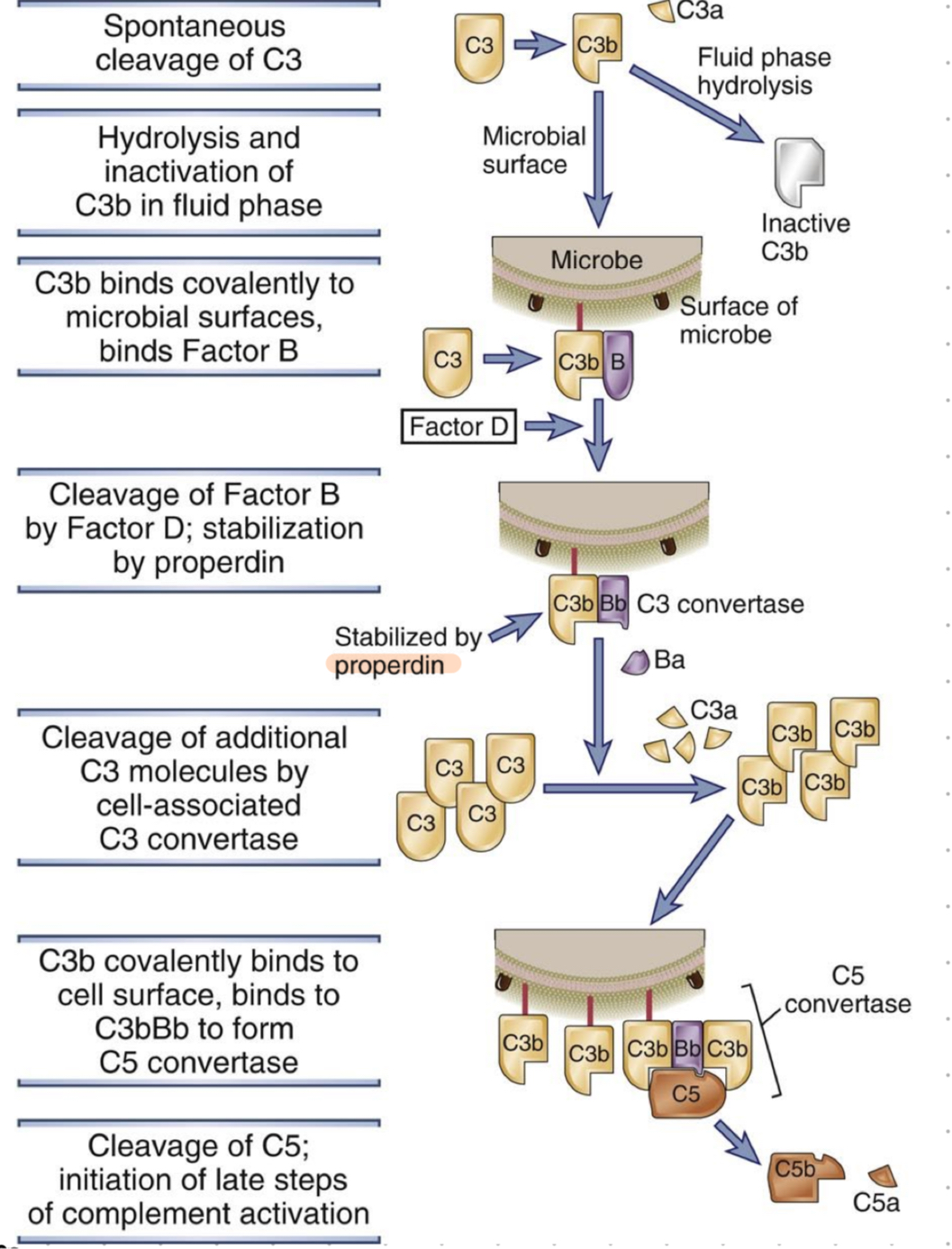
alternative pathway - step by step
spontaneous cleave of C3 (in blood)
inactivation of C3b OR binding to microbial surface
factor B binds, creates C3bB complex on microbe surface
cleavage of factor B by factor D, stabilisation by properdin
CrbBb (aka C3 convertase) cleaves additional C3 molecules (aka opsonisation)
C3b binds covalently to cell surface, binds also to C3bBb to form C5 convertase
cleavage of C5 → late steps of activation
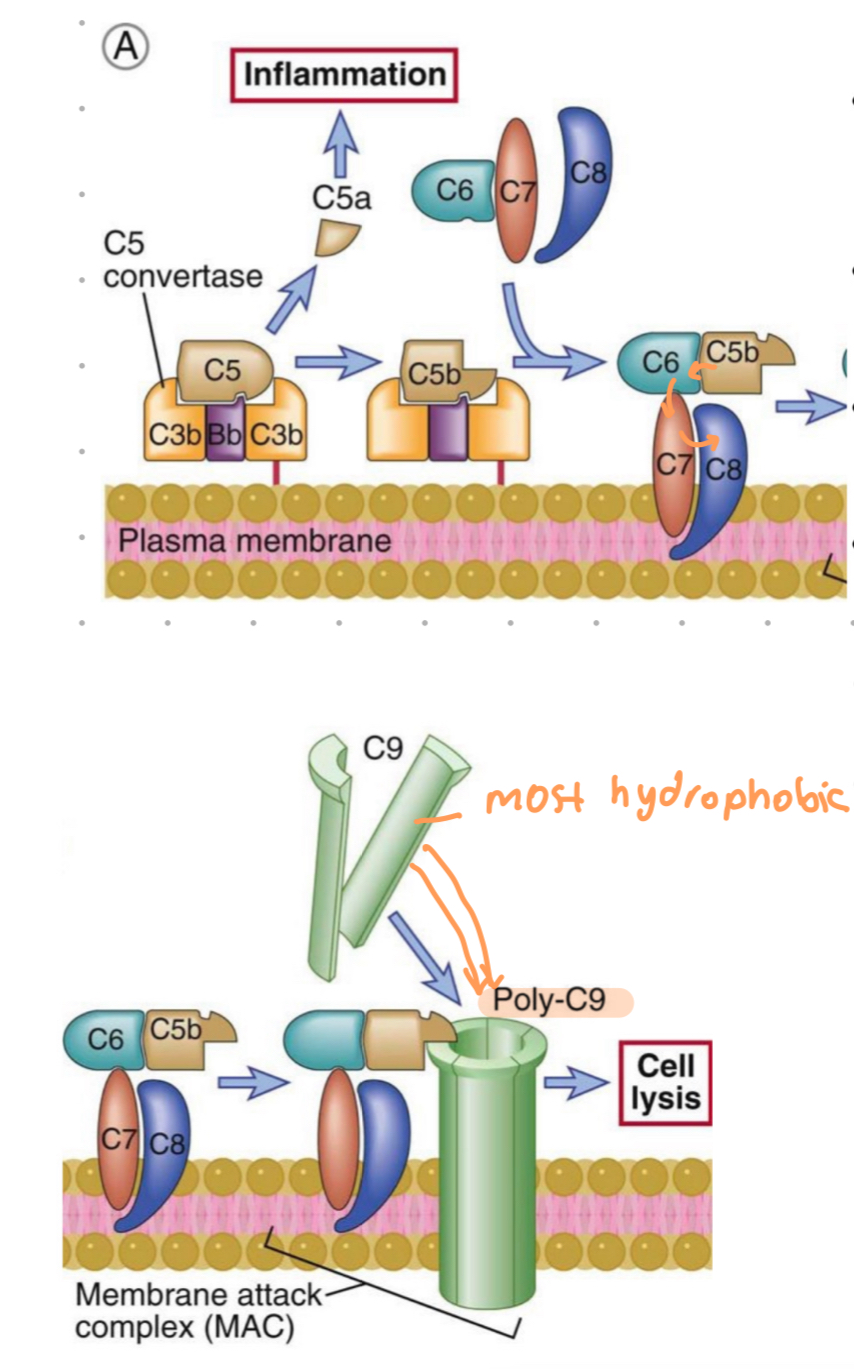
late steps of complement activation
convergence of all 3 pathways initiated by C5 binding to C5 convertase generated in all pathways
cleavage of C5
generation of C5a and C5b
C5b fragment remains bound to CP deposited on cell surface
C5b
transiently maintains a conformation capable of binding the next cascade proteins - C6 and C7
→ C5b,6,7 complex
C5b,6,7 complex
hydrophobic complex, insets into the lipid bilayer of cell membranes
becomes a high affinity receptor for the C8 molecule
C5b,6,7,8 complex
limited ability to lose cells
C9 is a serum protein that polymerises at site of bound C5b-8 complex to form MAC
C9
serum protein that polymerizes at site of the bound C5-8 complex, forming the membrane attack complex pores
MAC pores
pore in PM which are about 100A in diameter
form channels that allow free movement of water and ions
water entry = osmotic swelling + rupture of cell
aka osmotic lysis of bacteria
late steps - complement
C5 from all pathways cleaved
C5b binds C6 and C7
insertion of C5b,6,7 complex into PM
C8 binds, forms C5b-8 complex
C9 polymerizes at site of complex, forming MAC
C3 deficiency
associated w. frequent serious pyogenic bacterial infections
may be fatal
illustrates central role of C3, importance in opsonisation, enhanced phagocytosis, removal of bacteria
C2 deficiency
most common complement deficiency
resemble autoimmune disease systemic lupus erythematosus
CR1 on phagocytes
aka - type 1 complement receptor
recognises C3b opsonized microbes, initiates killing of the microbe
anaphylatoxins
potent stimulators of inflammation
C3a and C5a
mammalian regulatory proteins
mammalian cells express regulatory proteins that inhibit complement activation
prevents complement mediated damage to host
microbes lack regulatory proteins and therefore susceptible to complement
decay-accelerating factor (DAF)
membrane protein that disrupts the binding of factor B to C3b, or binding of C4b2a to C3b
halts complement activation in both alternative and classical pathways
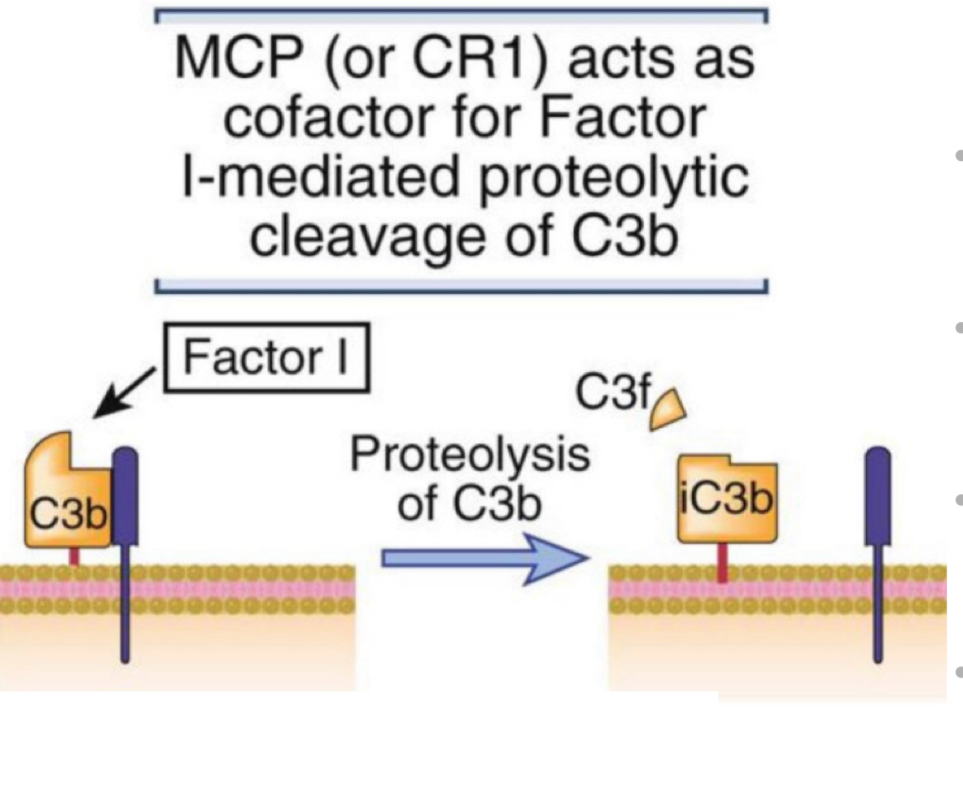
membrane cofactor protein (MCP)
cofactor for proteolytic breakdown of C3b into inactive iC3b
mediated by plasma enzyme factor 14
membrane bound complement inhibitors
regulatory proteins on mammalian cells
decay accelerating factor (DAF)
membrane cofactor protein (MCP)
C1 inhibitor(C1 INH)
a soluble inhibitor
prevents C1 complex assembly, blocking classical pathway
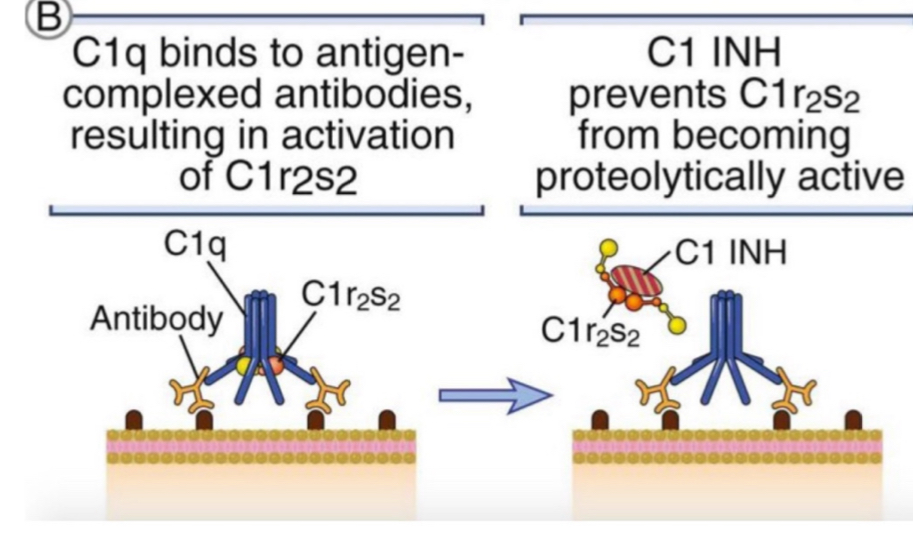
C1 INH deficiency
leads to excessive C1 activation and production of vasoactive protein fragments → leakage of fluid (edema) in the larynx and other tissues
DAF deficiency
results in unregulated complement activation on erythrocytes, lysis of erthrocytes