BIOL1107 Exam 2
1/81
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
82 Terms
Describe that major experiments that led to the identification of DNA as the hereditary molecule:
Griffith: his experiment on mice showed that virulent bacteria could be passed down through DNA (as opposed to proteins) which showed DNA was the hereditary molecule
Watson & Crick (1953): worked together to create double-helix model from Franklin’s ‘Photo 51’, eventually awarded Nobel Prize for discovery
Franklin/Wilkins: took x-ray crystallography photographs of DNA — Rosalind Franklin discovered helical structure but Wilkins gave information to Watson and Crick
Why did Watson and Crick originally think DNA was a triple helix? What made them change their mind?
they originally thought DNA was a triple helix with bases on the outside because the bases are the most important part and should be on the exterior
‘Photo 51’ made them revise their model
Even before DNA structure was solved, what properties of DNA were known?
DNA was able to store information
DNA could be replicated and passed on from generation to generation
DNA allows for changes and thus evolution to occur
Explain how the sequence of a molecule of DNA, made up of only four possible nucleotides, can encode the enormous amount of genetic information stored in the chromosomes of living organisms.
four nucleotide monomers: adenine & thymine / cytosine & guanine (RNA; adenine & uracil / cytosine & guanine)
because the four nucleotides can be in any order/sequence
any base strand of DNA can be followed by any other base (or the same base) which gives rise to enormous genetic diversity
What is the Central Dogma? Describe the usual flow of genetic information in the cell.
DNA to RNA to protein
What is transcription? What are the 3 steps?
occurs in the nucleus of eukaryotes and in the cytoplasm of prokaryotes
DNA to mRNA: template strand of DNA is transcribed to create a strand of RNA
in simple terms: the copying of genetic info from DNA
Step 1 Initiation:
RNA polymerase (enzyme) binds to promotor sequence of DNA marking the starting point of transcription
in eukaryotes, transcription factors are needed to begin initiation
it unwinds DNA helix to expose single DNA strands (template & coding strand)
region is called the transcription bubble
Step 2 Elongation:
RNA polymerase reads DNA template from 3’ to 5’ and begins to transcribe RNA strand in the complimentary, 5’ to 3’ direction
complimentary nucleotides are added to the growing mRNA chain
as RNA polymerase progresses, it released the DNA template strand
Step 3 Termination:
RNA polymerase reaches the transcription stop site and falls off template strand, a strand of newly synthesized RNA is released into the cytoplasm where it will undergo translation, and the DNA helix reforms
Additional Note: in eukaryotes, RNA undergoes further processing including capping and splicing before it is mature and ready to be translated into protein
What is translation? What are the 3 steps?
occurs in ribosome
RNA to protein: RNA strand is translated into a sequence of amino acids during protein synthesis
tRNAs carry specific amino acids and have complementary anticodons that base-pair with the codons of mRNA
Step 1 Initiation:
small ribosomal subunit attaches to the mRNA at the start codon (AUG) via initiation factors
initiator tRNA carries the amino acid Methionine (MET) to bind to the start codon
large ribosomal subunit joins
Step 2 Elongation: (aka polymerization)
ribosome moves along mRNA in 5’ to 3’ direction
tRNAs carry amino acids to the ribosome
the tRNA’s anticodon base pairs with the mRNA codon
amino acids form peptide bonds between each other, forming a growing polypeptide chain
tRNAs moves throughout large subunit of ribosome (E,P,A sites)
Step 3 Termination:
ribosome recognizes a stop codon (UGA/UAG/UAA) and a release factor binds to ribosome
ribosome releases completed polypeptide chain
polypeptide then folds into a specific protein depending on specific structure of amino acids
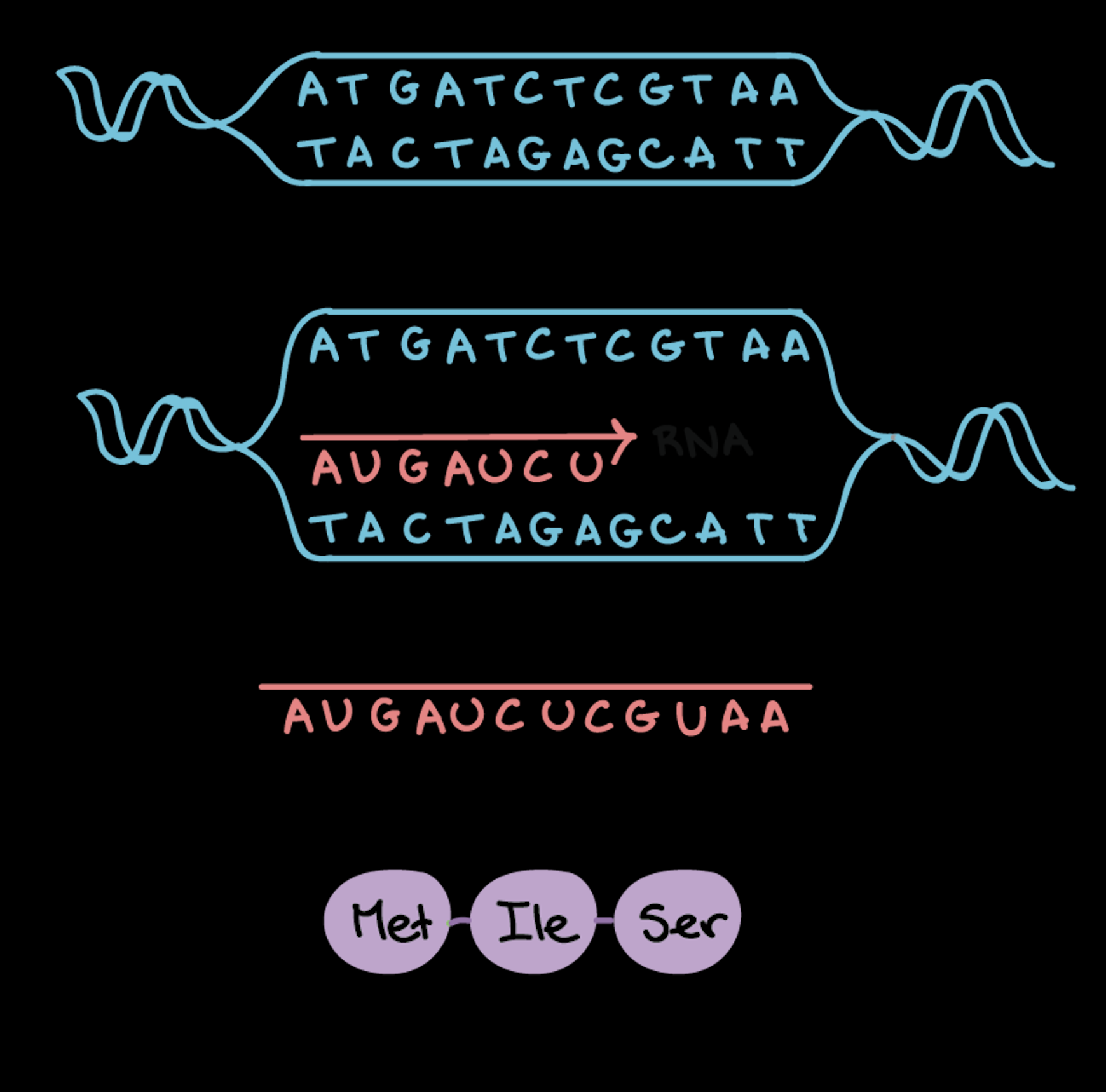
Compare and contrast transcription and translation:
Similarities
both are essential processes in the central dogma
both involve synthesis of molecules that affect biological processes (i.e., mRNA and proteins)
Differences:
the products are different as transcription produces mRNA and translation produces a polypeptide chain
mRNA carries genetic information from DNA to the ribosome
polypeptide chain eventually forms a protein
the enzymes are different as transcription is catalyzed by RNA polymerase and translation involves ribosomes, tRNAs, and aaRSs
aaRSs is aminoacyl-tRNA synthetases: they are just enzymes that match an amino acid to its corresponding anticodon of tRNA
they are specific for each amino acid
the location is different as transcription occurs in the nucleus (in eukaryotes) or cytoplasm (in prokaryotes) while translation always takes place in the ribosomes (which are located in the cytoplasm)
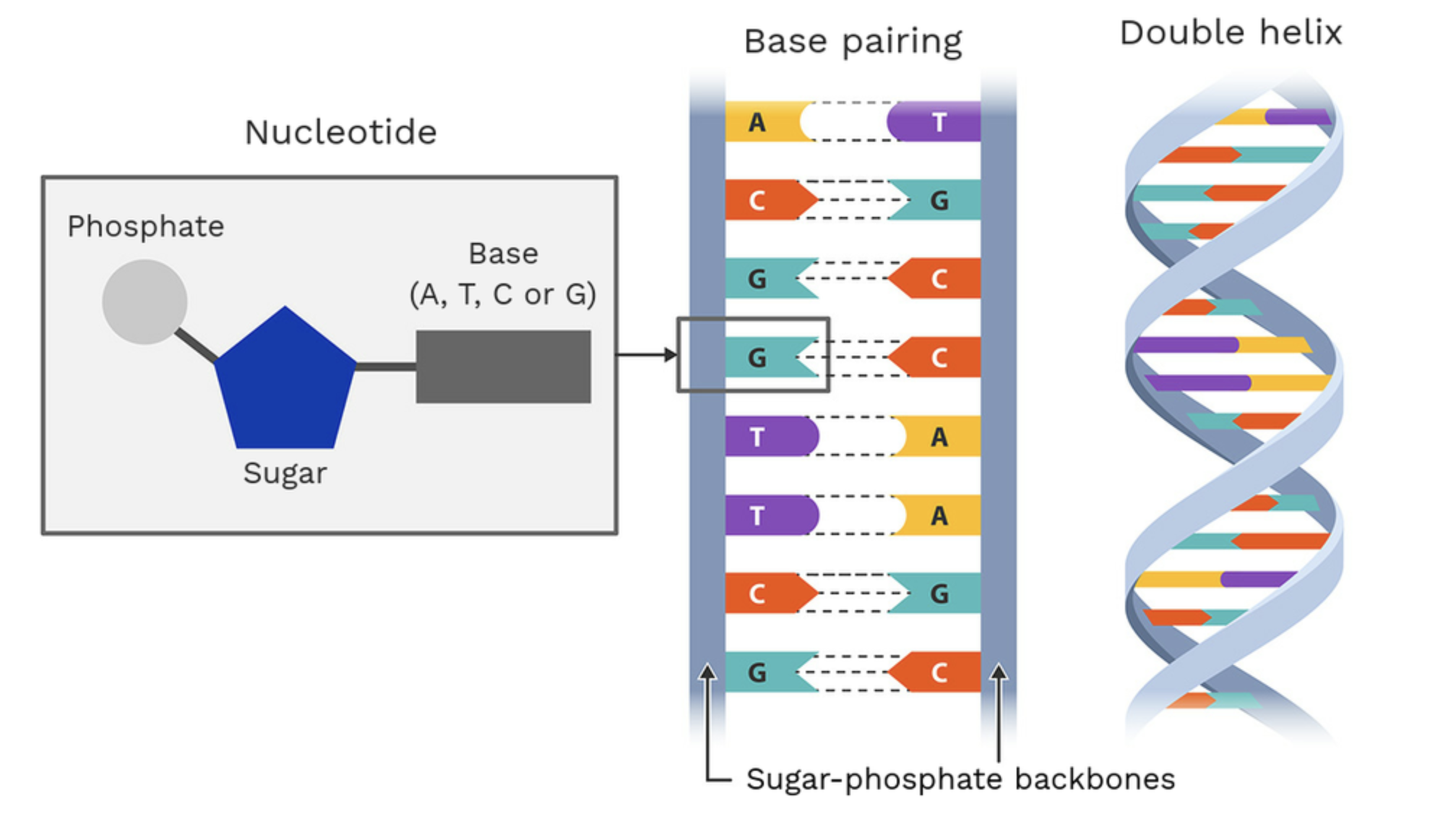
What is DNA? Describe its structure:
DNA is a double-stranded macromolecule that carries genetic information. It is composed of nucleotides which are the building blocks of DNA. Each nucleotide consists of:
a 5-carbon sugar known as deoxyribose that forms backbone of the DNA strand
a phosphate group: attached to the 5th carbon of the 5-carbon sugar & forms the sugar-phosphate backbone
a nitrogenous base (four types: A, T, G, C) which are attached to the 1st carbon of the 5-carbon sugar
double helix formation; two strands run antiparallel to each other (in opposite direction) due to orientation of sugar-phosphate backbone
wound tightly around each other for efficient storage of genetic information
sugar-phosphate backbone with bases on interior
complimentary base pairing via hydrogen bonds
Why is DNA antiparallel? What would happen if DNA strands were not antiparallel?
refers to how the two DNA strands run in opposite directions
one strand is built in the 5’ to 3’ direction and the other is in 3’ to 5’ direction
strands are antiparallel to ensure nitrogenous bases pair correctly which allows for accurate DNA replication and transcription
antiparallel due to hydrogen bonding
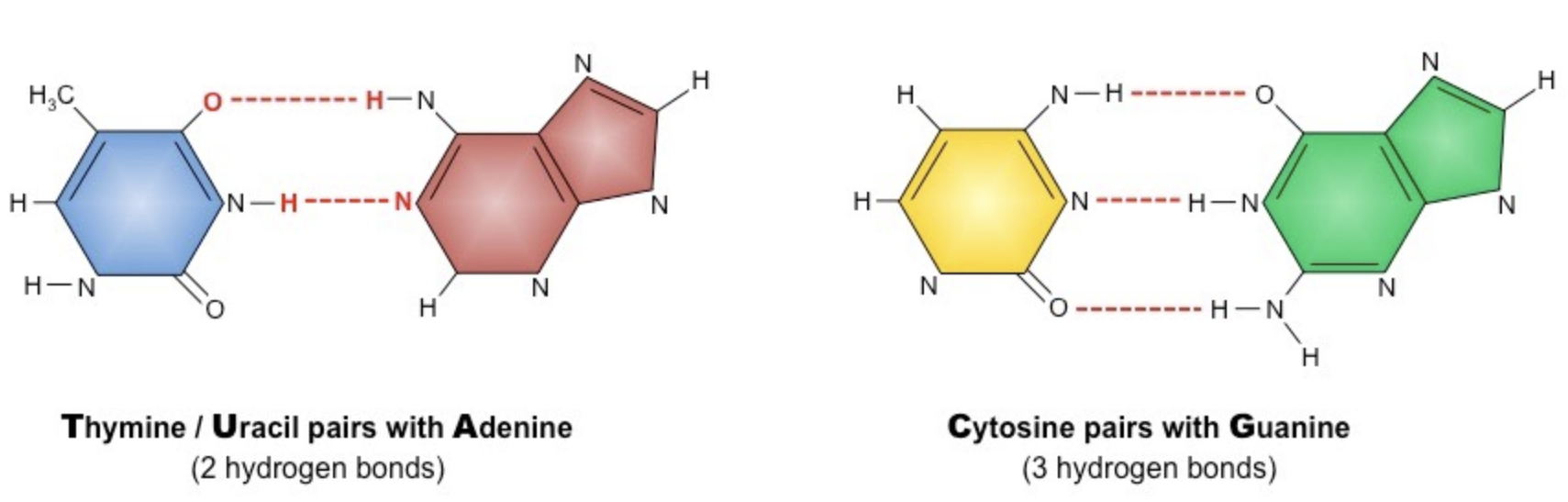
Adenine & Thymine form two hydrogen bonds (A&U in RNA)
Guanine & Cytosine form three hydrogen bonds
if strands were not antiparallel, base pairing would not occur correctly and the genetic information would not be accurately replicated or transcribed
Compare & Contrast the 5’ and 3’ end of a DNA strand:
on a single strand of DNA
the 5’ end terminates with a phosphate group which is attached to the 5’ carbon of the 5-carbon sugar
considered the “upstream” end
starting point for the synthesis of a new DNA strand during replication
where nucleotides attach
the 3’ end terminates with a hydroxyl group (-OH) which is attached to the 3’ carbon of the 5-carbon sugar
considered the “downstream” end
site where new nucleotides are added during replication
extends RNA chain
Comparison:
both are part of the sugar-phosphate backbone of DNA strand
both involve phosphodiester bonds which link adjacent nucleotides in DNA strand
Contrast:
5’ end terminates with phosphate group while 3’ end terminates with hydroxyl group
5’ end is where initiation begins during DNA synthesis while 3’ end is the site of elongation (in which nucleotides are added)
When a mixture of debris from killed virulent bacteria and live non-virulent bacteria are injected into mice:
A) the mice are killed because the virulent bacteria are revived
B) the mice are killed because the nonvirulent bacteria are transformed into virulent bacteria
C) the mice survive because none of the living bacteria are virulent
D) the mice survive for a period of a few weeks and then die from unrelated illness
E) half the mice are killed and half survive
B) the mice are killed because the nonvirulent bacteria are transformed into virulent bacteria
Which statement is true of DNA?
A) it is used by the ribosome for translation
B) successive nucleotides in a strand are connected by hydrogen bonds
C) a phosphate group in a nucleotide is attached to the 3’ carbon in ribose
D) the percentage of the purine A always equals the percentage of the purine G
E) a purine always forms a complimentary base pair with a pyrimidine
E) a purine always forms a complimentary base pair with a pyrimidine
What are the 4 key differences between the structure of DNA and RNA?
sugar backbone:
in DNA sugar backbone is deoxyribose
in RNA sugar backbone is ribose
RNA has hydroxyl group (-OH) on 2’ carbon while DNA only has H on 2’ carbon
DE-OXY meaning NO OXYGEN (i.e., just hydrogen for DNA)
bases:
in DNA A pairs with T and C pairs with G
in RNA A pairs with U and C pairs with G
U instead of T
stability:
DNA is more stable than RNA
RNA is more susceptible to degradation because the hydroxyl group makes the molecule more chemically reactive
single vs double stranded:
DNA is double stranded with two antiparallel strands
RNA is single stranded
DNA is primarily responsible for storing genetic information
RNA takes part in transcription (copying genetic info from DNA) and translation (making proteins based on genetic info)
Describe how a molecule of RNA is synthesized using a DNA molecule as a template:
An exon is…
A) a transfer RNA (tRNA) that binds to the codon
B) a protein that is modified post-translationally
C) RNA that is removed during the processing of an RNA molecule and remains inside the nucleus
D) part of an intact, mature mRNA that leaves the nucleus
E) a series of amino acids at the end of a new polypeptide that directs transcription to the ER
D) part of an intact, mature mRNA that leaves the nucleus
If you made a change in the promotor sequence in the DNA that inactivates the promoter, what would happen at the RNA level?
A) nothing because the RNA would be made as usual
B) the RNA polymerase would not be able to recognize and bind the DNA, so no RNA would be made
C) the mutation of DNA would be carried through to the RNA sequence
D) DNA helicase would not be able to recognize and bind the DNA so the RNA would not be made
B) the RNA polymerase would not be able to recognize and bind the DNA, so no RNA would be made
In eukaryotes, where do activator proteins bind?
A) RNA polymerase
B) terminators
C) DNA polymerase
D) promoters
E) enhancers
E) enhancers
Which of the actions would affect the secondary structure of a protein? Select all that apply.
A) breaking hydrogen bonds between amino acids
B) breaking ionic between amino acids
C) changing sequence of amino acids
D) disruption of interactions between two different polypeptide chains
A) breaking hydrogen bonds between amino acids
C) changing sequence of amino acids
Which force can contribute to a protein’s tertiary structure?
A) ionic bonding
B) Van der Waal's forces
C) covalent bonding
D) hydrogen bonding
E) all of these choices are correct
E) all of these choices are correct
Which statements are the steps in the elongation phase of translation?
A) introduction of a charges tRNA into an unoccupied A site of a ribosome
B) ejection of an uncharged tRNA from the E site of the ribosome
C) peptide bond formation
D) movement of the ribosome three bases closer to the 3’ end of mRNA
E) all of these choices are correct
E) all of these choices are correct
Which step occurs in the P site of the ribosome during translation?
A) an incoming charged tRNA binds to this site
B) tRNA carrying growing polypeptide moves to the site as the ribosome slides to the next codon
C) an uncharged tRNA is ejected from this site as the ribosome slides to the next codon
D) none of the other answer options is correct
B) tRNA carrying growing polypeptide moves to the site as the ribosome slides to the next codon
Which of the statements describes catabolic reactions?
A) they are exergonic and have a positive change in free energy
B) they are endergonic and have a negative change in free energy
C) they are exergonic and have a negative change in free energy
D) they are endergonic and have a positive change in free energy
C) they are exergonic and have a negative change in free energy
Which of the reactions is most likely to be exergonic?
A) the digestion of protein from food into amino acids
B) the replication of DNA from free nucleotides
C) the formation of cellulose from individual glucose molecules
D) the synthesis of a phospholipid from glycerol and fatty acids
A) the digestion of protein from food into amino acids
Which of the statements is true of allosteric inhibitors of an enzyme?
A) allosteric inhibitors decrease enzyme activity
B) allosteric inhibitors bind to the active site of the enzyme
C) allosteric inhibitors are structurally similar to the normal substrate of an enzyme
D) allosteric inhibitors increase the rate of enzyme activity
A) allosteric inhibitors decrease enzyme activity
Production of NADH by glycolysis requires an input of of the oxidized molecule NAD+. Where does this supply of NAD+ come from in the absence of oxygen?
A) it is regenerated by donating electrons to the electron transport chain
B) it is regenerated by reducing pyruvate to ethanol or lactic acid
C) it is transferred from the citric acid cycle, which is not functioning due to lack of oxygen
D) there is a ready supply of NAD+ from the cytoplasm
B) it is regenerated by reducing pyruvate to ethanol or lactic acid
During the metabolism of glucose (the cellular fuel molecule) by the process of aerobic cellular respiration, glucose is ___ via ___ reactions.
A) oxidized; redox
b) reduced; endergonic
C) oxidized; hydrolysis
D) reduced; dehydration
E) reduced; oxidation
A) oxidized; redox
Which summary sequence correctly tracks electrons through the overall process of aerobic cellular respiration?
A) glucose → electron carriers → electron transport chain → water
B) glucose → pyruvate → acetyl-CoA → CO2
C) glucose → pyruvate → acetyl-CoA → CO2 → ATP
D) NAD+ and FAD → NADH and FADH2 → ATP
A) glucose → electron carriers → electron transport chain → water
Certain complexes of the mitochondrial electron transport chain pump protons. Protons are pumped across the ___ mitochondrial membrane, from the ___ to the ___ .
A) inner; matrix; intermembrane space
B) outer; cytoplasm; intermembrane space
C) outer; intermembrane space; cytoplasm
D) inner; intermembrane space; matrix
D) inner; intermembrane space; matrix
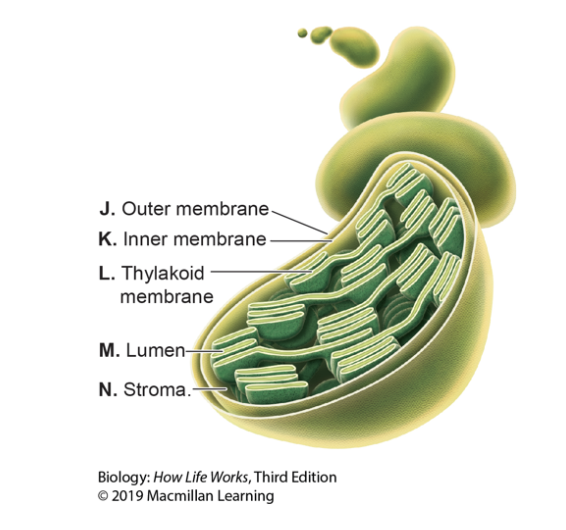
Refer to the image. In which lettered location in the figure would you find electrons moving between large chlorophyll-containing complexes?
A) J
B) K
C) L
D) M
E) N
C) L
During photosynthesis, ___ is reduced to ___ .
A) CO2; O2
B) H2O; CO2
C) O2; H2O
D) sugar; O2
E) CO2; sugar
E) CO2; sugar
What is a phosphodiester bond? How does it relate to DNA/RNA?
bonds in the sugar phosphate backbone of DNA which link nucleotides together
the covalent bonds of the 3rd carbon of one sugar to the fifth carbon of the next sugar
stabilizes structure of DNA & RNA
How do bases pair? (hint: a type of bonding) What is its purpose? How does base-pairing differ in DNA vs RNA?
nitrogenous bases pair via hydrogen bonding
Adenine always pair with Thymine and Cytosine always pairs with Guanine
in RNA, the Thymine is switched to Uracil
base-pairing rule: complimentary base pairing ensures that each strand of DNA serves as a template for the synthesis of its new complementary strand
purpose is to ensure that genetic information is accurately passed down from one generation to the next
What are regulatory sequence/elements in DNA and what is their purpose?
present in the transcription process in eukaryotes
regulatory elements include promoters and enhancers; influence rate of transcription initiation
control when the gene starts and how the gene is expressed
crucial role in regulation of gene activity
What is genetic redundancy in terms of DNA?
it means that the genetic code is degenerate, meaning that two or more genes are performing the same function
i.e, multiple codons can code for the same amino acid
this corrects error and makes organisms more resistant to mutations because a mutation in one single gene will have a smaller effect on the organism since multiple different genes are performing the same function
What is the flow of genetic information in a cell?
DNA is transcribed into messenger RNA (mRNA) through transcription and then mRNA is translated into a protein through translation
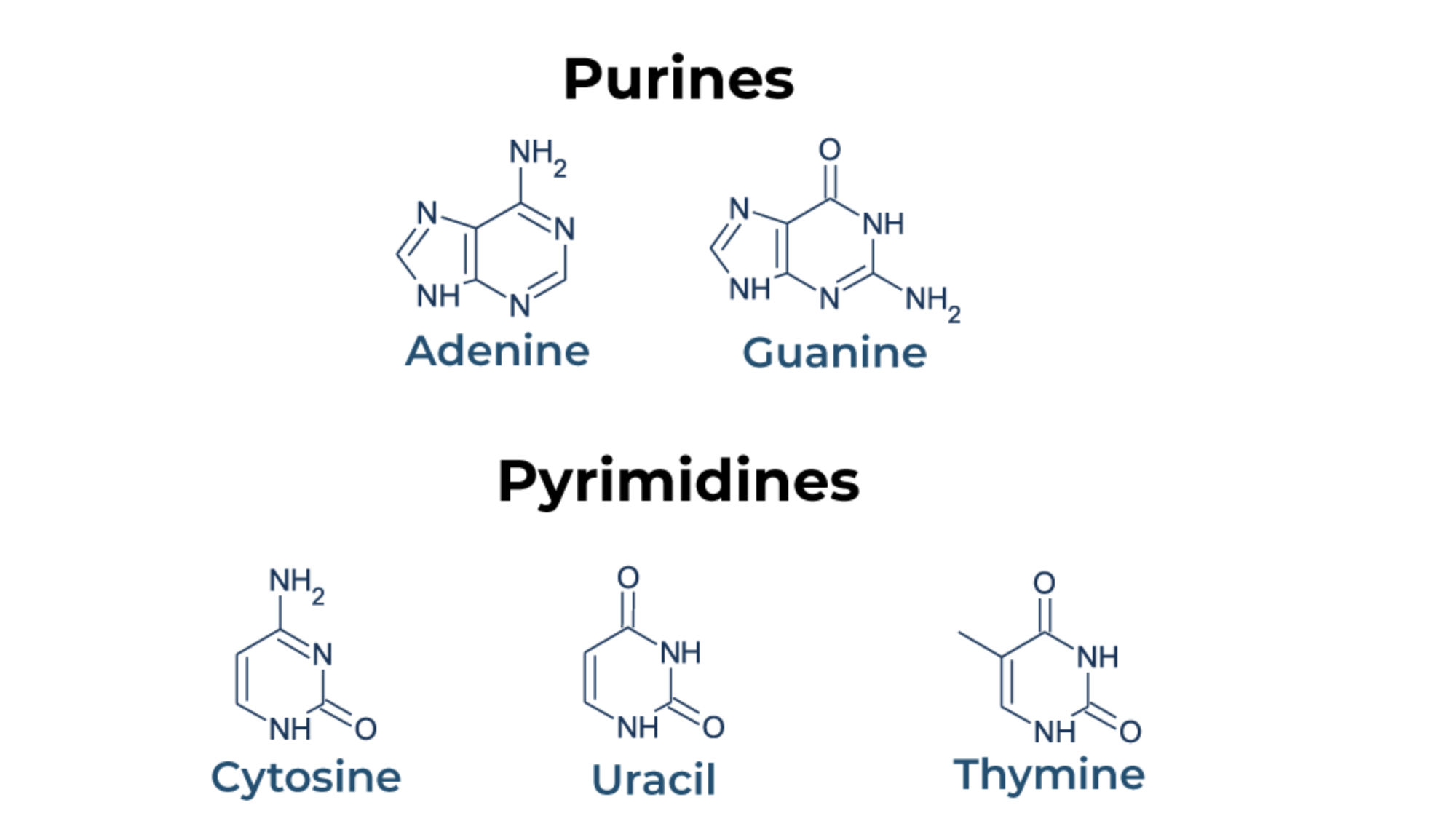
What are purine bases in DNA/RNA? What are the pyrimidine bases? What differentiates them?
purine bases are Adenine (A) and Guanine (G) because they have two carbon rings
mnemonic: Pure As Gold
Purines = Adenine and Guanine
pyrimidine bases are Thymine (T) and Cytosine (C) (and Uracil (U) in RNA) because they have a single carbon ring
Given a DNA template strand, you can predict the sequence of its complementary strand by applying the base-pairing rules. What is the complimentary strand if the template strand is AGTACCGG
the complimentary strand is TCATGGCC
What is RNA polymerase and what process is it involved in?
it is the enzyme responsible for transcription in which a new strand of mRNA is synthesized from a template strand of DNA
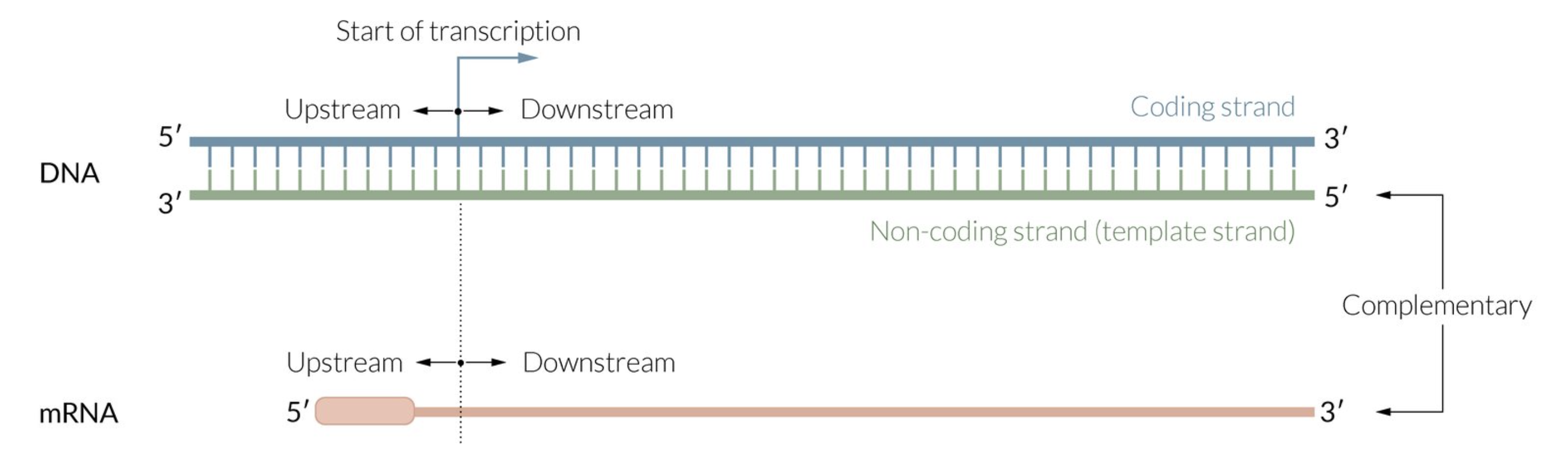
What is considered upstream vs downstream in DNA and RNA?
in DNA: double stranded
“upstream” is towards the 5’ end of the coding strand (top strand)
“downstream” is towards the 3’ end of the coding strand
coding strand is on top and non-coding strand (aka template strand) is on bottom
in RNA: single stranded
“upstream” is towards the 5’ end
“downstream” is towards the 3’ end
What is a promotor sequence? What does it do? What is the TATA box?
it is a specific DNA sequence that serves as the binding site for RNA polymerase which initiates the transcription process
located upstream (towards 5’ end) of transcribed gene
TATA box: a type of promotor sequence which specifies to other molecules where transcription begins
helps RNA polymerase identify the start site for transcription
What are transcription factors? What are activators/repressors?
they are proteins that bind to specific DNA sequences near the promoter and their purpose is to regulate transcription
activators: proteins that enhance initiation of transcription by helping RNA polymerase bind to the promotor
repressors: proteins that inhibit transcription process by blocking RNA polymerase from binding to the promoter
What is the template strand vs the coding strand of DNA?
template strand (aka non-coding) : serves as template for the synthesis of the complementary RNA strand
coding strand: complementary to the template strand and therefore has the same sequence as the newly synthesized RNA strand (except with Thymine (T) as opposed to Uracil (U) obviously since its DNA)
What is the structure and function of RNA?
single-stranded structure
complementary base pairing: RNA uses the same bases as DNA with the exception of Uracil (U) in place of Thymine (T)
allows RNA to base-pair with DNA during transcription & other molecules
enables translation and RNA folding
functional diversity: RNA has lots of functions
examples: mRNA carries genetic information and tRNA aiding in translation process/protein synthesis
What is the source of energy for polymerization/transcription of RNA from DNA?
nucleoside triphosphate (aka NTPs) are the building blocks of RNA and they provide energy for polymerization via hydrolysis
NTP incorporated into growing RNA chain, phosphate bonds are broken, releasing energy, energy is used to form phosphodiester bond between new nucleotide and existing chain
hydrolysis: breakdown of compound by addition of water
RNA processing in eukaryotes involve several crucial steps to produce mature, functional RNA molecules. What are the 3 steps?
5’ Capping: the 5’ cap is a modified guanine nucleotide that is added to the 5’ end of the pre-mRNA during transcription
protects RNA molecule from degradation
crucial role in translation — provides a site for ribosome to bind to the mRNA
facilitates export of mature mRNA from nucleus to cytoplasm where translation will take place
Splicing: the process by which non-coding introns are removed from the pre-mRNA and the coding extrons are joined together
achieved by the spliceosome which recognizes splice sites within pre-mRNA
important in production of function mature mRNA because it ensures that only the coding info remains
provides yet another level of regulation — alternative splicing can produce different proteins from one single gene
Poly-A-Tail: a tail of Adenine nucleotides are added to the 3’ end of the pre-mRNA
provides stability the mRNA by protecting it from degradation
helps export mRNA from the nucleus to the cytoplasm
recognition signal for ribosome during translation initiation
What is alternative splicing? What is the evolutionary significance?
it is a process in which different combinations of exons can be joined together resulting in the production of multiple mRNA from one single gene
evolutionary significance:
expands diversity of proteins within an organism
increases functional diversity of genes without increasing the number of genes in an organisms genome (important in complex organisms with a limited number of genes i.e, humans)
allows organisms to adapt to changing environment & prevent virus
***biological complexity & adaptability of eukaryotic organisms!!!
What are the types of non-coding RNA? What are their functions?
Ribosomal RNA (rRNA) and Transfer RNA (tRNA) both do not encode proteins but carry out other crucial roles
rRNA: key component of ribosomes (made up of protein and rRNA), structural framework & catalytic sites of ribosome
rRNA interacts with tRNA and mRNA during translation: ensures accurate pairing of codons on mRNA with anticodons on tRNA
tRNA: interprets genetic code during protein synthesis, carries amino acids to ribosome where they are incorporated into growing polypeptide chain
each tRNA is specific to a particular amino acid (one end binds to amino acid and the other carries complementary anticodon)
anticodons ensure that the correct amino acid is added to growing chain
both are essential in protein synthesis!
What is the structure of protein? (hint: what are the monomers) How is a peptide bond formed? How does the R-group affect protein folding/shape?
proteins are made up of amino acid and each amino acid is linked by a peptide bond
amino acids consist of:
an amino group (NH2)
a carboxyl group (COOH)
and a variable R-group/side chain
peptide bonds are formed through dehydration synthesis; small molecules are assembled into larger molecules by removing water
the carboxyl group (COOH) of one amino acid bonds to the amino group (NH2) of another amino acid form the covalent bond known as the peptide bond
R-groups
polar R-groups are hydrophilic and therefore interact with water molecules on the exterior of the protein
non-polar R-groups are hydrophobic and cluster together in the protein’s interior away from water
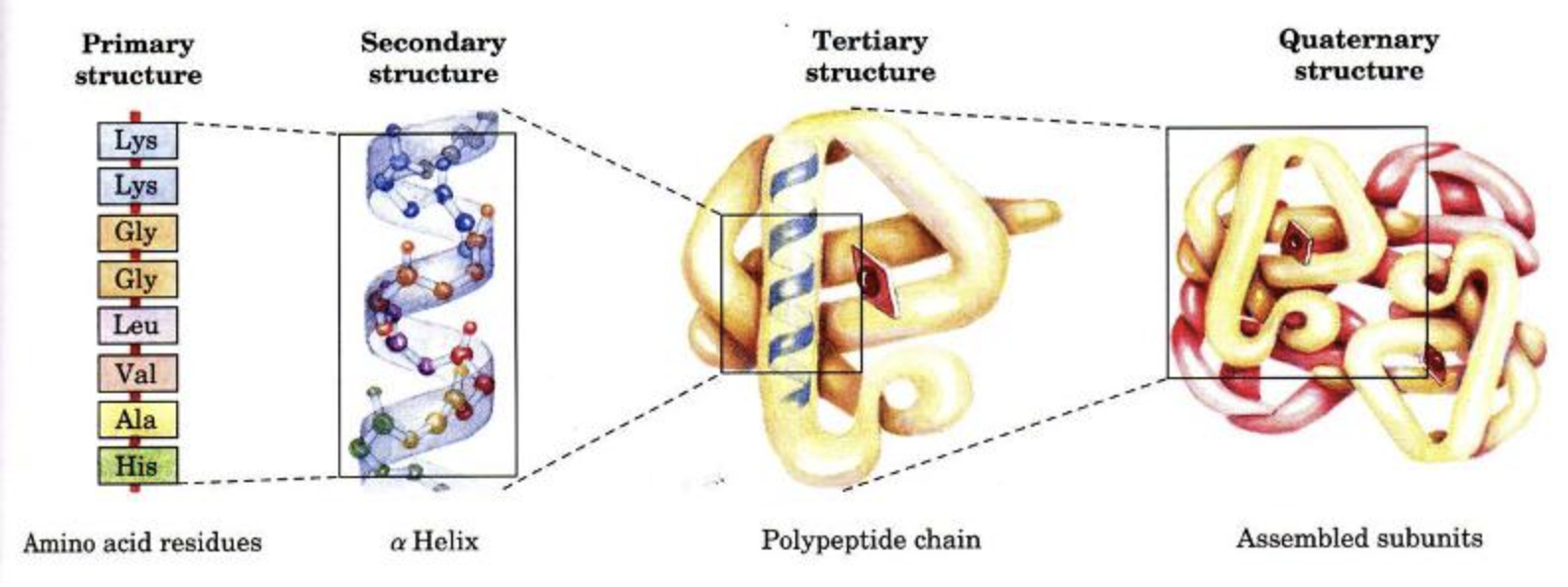
What are a protein’s four structural levels? How do peptide bonds, hydrogen bonds, ionic bonds, disulfide bridges, and non-covalent interactions (i.e., van der Waals forces & hydrophobic effect) define these levels?
four structures of protein:
primary: sequence of amino acids linked by peptide bonds
secondary: stabilized by hydrogen bonds between amino acids
alpha helices & beta sheets)
tertiary: 3-D structure of polypeptide chain maintained by disulfide bridges, hydrogen bonds, ionic bonds, & hydrophobic interactions
quaternary structure: interaction between multiple polypeptide chains within a protein
How do amino acids affect protein folding and function? How do mutated amino acids affect proteins?
sequence of AA (primary structure) affect how protein will fold
certain sequences promote formation of alpha helices and beta sheets (secondary structure) which then lead to overall 3-D folding of protein
protein’s folding allows protein to interact with other molecules (i.e., substrates/other proteins)
mutations alter AA sequence which can lead to misfolding or loss of function
mutation could introduce AA with different R-group which disrupts folding pattern & function
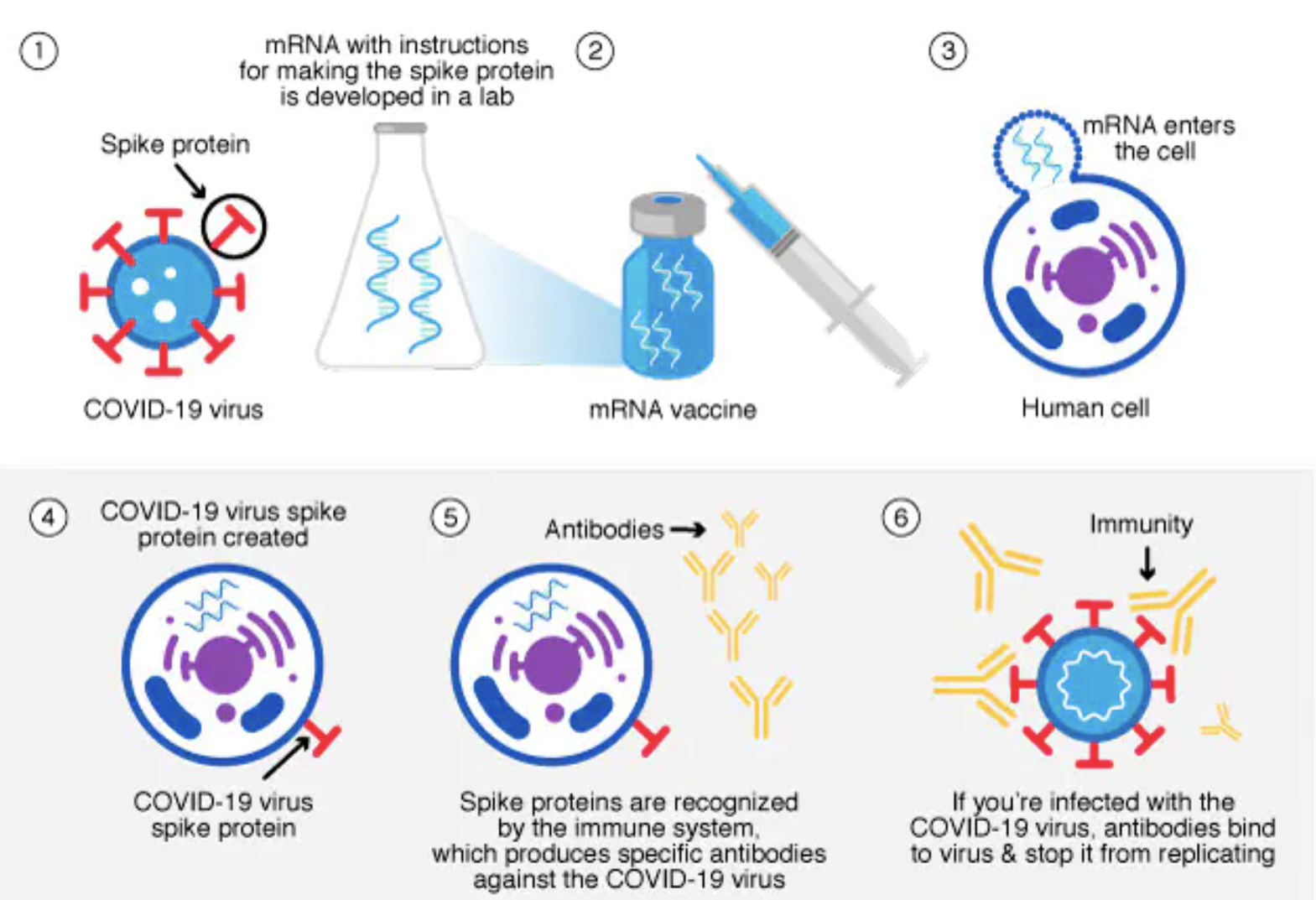
How does mRNA work in Covid vaccines?
covid vaccines use the mRNA of the virus to instruct cells to produce a spike protein which triggers an immune response and trains the body to recognize/fight virus
How many amino acids are there? How many codons? What does this say about genetic code?
there are 20 amino acids and 64 codons (sequence of 3 nucleotide bases)
this means that multiple codons code for the same amino acid (they are “degenerate”)
What is genetic code? What are the important codons?
it is the set of rules that dictate the correspondence between codons in mRNA and the specific amino acids they code for
important codons:
start codon — AUG which codes for methionine amino acid
stop codons — (UAG, UGA, UAA) which signal the end of translation
What are the ways in which organisms obtain energy and carbon from the environment?
autotrophs (self feeders): produce their own energy/carbon
i.e plants/photosynthesizes use energy from sunlight to convert CO2 into glucose
heterotrophs (other feeders): obtains energy/carbon from consuming other organisms (organic molecules like sugars/proteins)
What is catabolism? What is anabolism?
catabolism: breakdown of complex molecules which releases energy
i.e., cellular respiration: glucose is broken down to release usable energy for the cell
exergonic: energy goes OUT
spontaneous = - ΔG (free energy)
the CAT knocks the glass off the counter and breaks it
anabolism: synthesis of complex molecules which requires energy
i.e., protein synthesis: proteins are synthesizes from amino acids which is a process that requires energy
endergonic: energy goes IN
non-spontaneous = + ΔG (free energy)
ANA puts together the glass which requires energy
What are potential and kinetic energy?
potential energy: stored energy
ball at the top of the stairs
kinetic: energy of motion
moving car
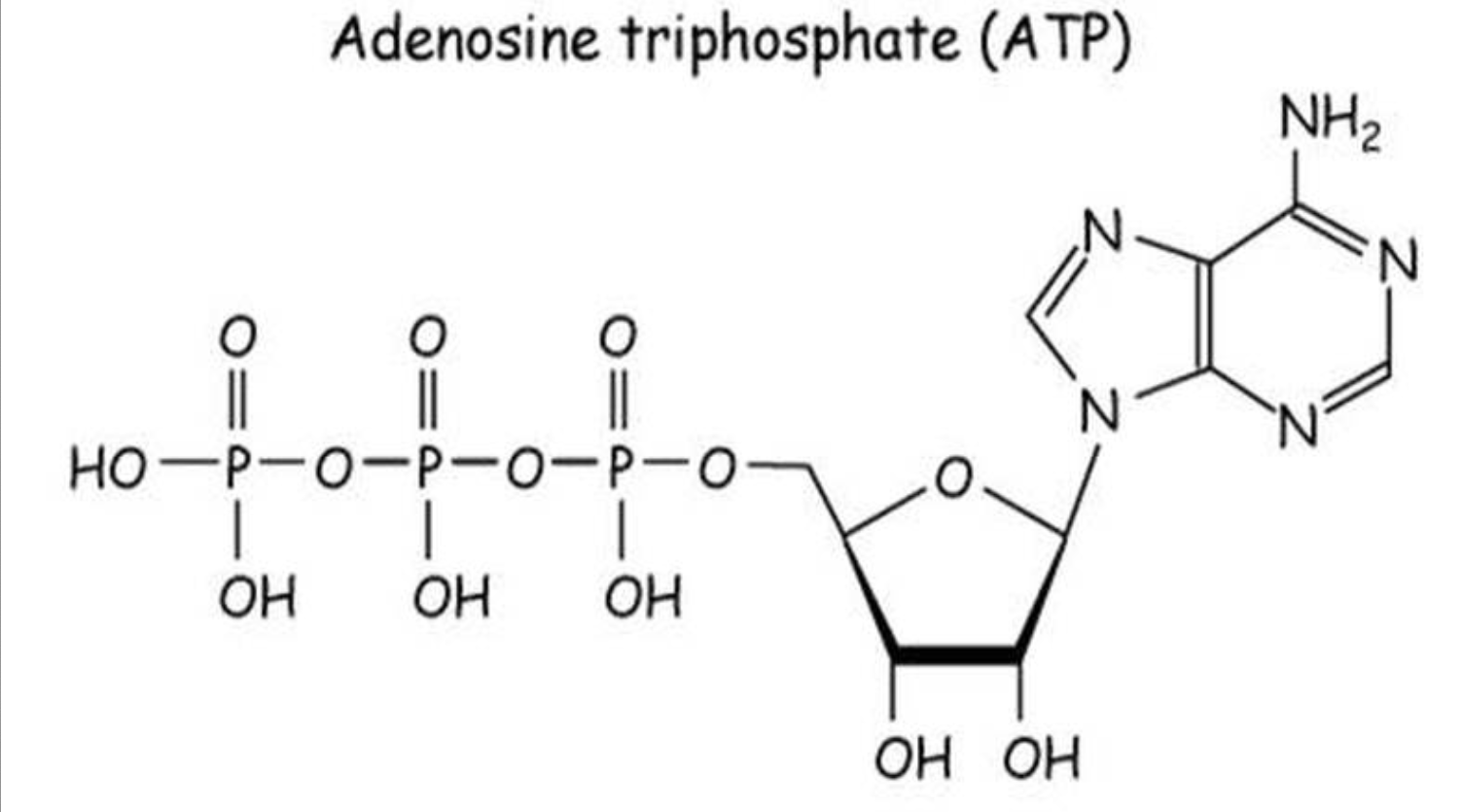
What is the structure of ATP? What bonds are broken during hydrolysis?
ATP = adenine triphosphate
structure:
adenine abse
ribose sugar
3 phosphate groups (hence TRI-phosphate)
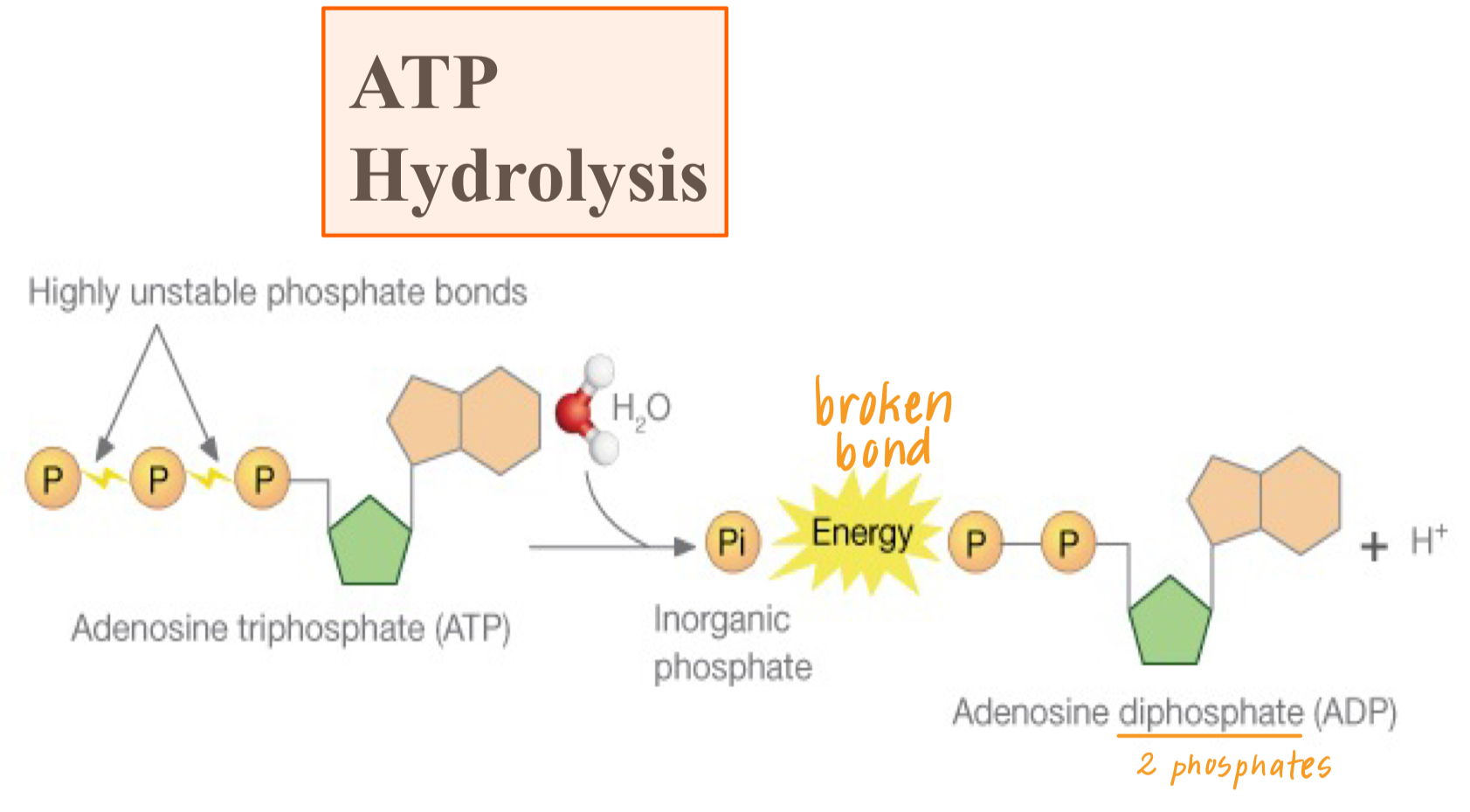
hydrolysis: converts ATP —> ADP and a phosphate group
ΔG (free energy) is negative/decreasing, so the reaction is spontaneous
water (H2O) is added during hydrolysis and the terminal phosphate group is broken off, releasing energy
What are the First & Second Law of Thermodynamics? How do they relate to chemical reactions?
First Law: energy cannot be created nor destroyed, only converted
Second Law: energy transformations always end in an increase in disorder in the universe
relates to chemical reactions because it shows how energy able to do work decreases every time energy changes forms
When is a reaction spontaneous?
a reaction is spontaneous when they result in a decrease in Gibbs free energy (-∆G)
What is energy coupling? How can ATP hydrolysis drive non-spontaneous reactions in a cell?
energy coupling: when energy released by an exergonic reaction is used to power/drive an endergonic reaction
exergonic: releases energy (ATP)
endergonic: requires an input of ATP
hydrolysis of ATP: when ATP is broken down into ADP and Pi by the addition of water which releases energy
this energy drives non-spontaneous/endergonic reactions in a cell
i.e glycolysis: free energy of ATP hydrolysis fuels glucose phosphorylation
What are the 3 characteristics of enzymes? How do enzymes permit chemical reactions to occur in cells?
enzyme characteristics:
specificity: enzymes are highly specific for their substrates
catalytic activity: speed up chemical reactions without being consumed
regulation: regulated to control metabolic pathways
enzymes can be regulated by activators/inhibitors that either increase or reduce their activity
enzymes & chemical reactions: they are catalysts that speed up reactions by helping to lower the activation energy needed to start a reaction
lower activation energy = faster reaction
each enzyme has an active site where another molecule, called the substrate, fits
enzymes end in “ase”
substrate: a molecule acted upon by an enzyme which is converted to a product
i.e., carbohydrates like glucose, sucrose, starch
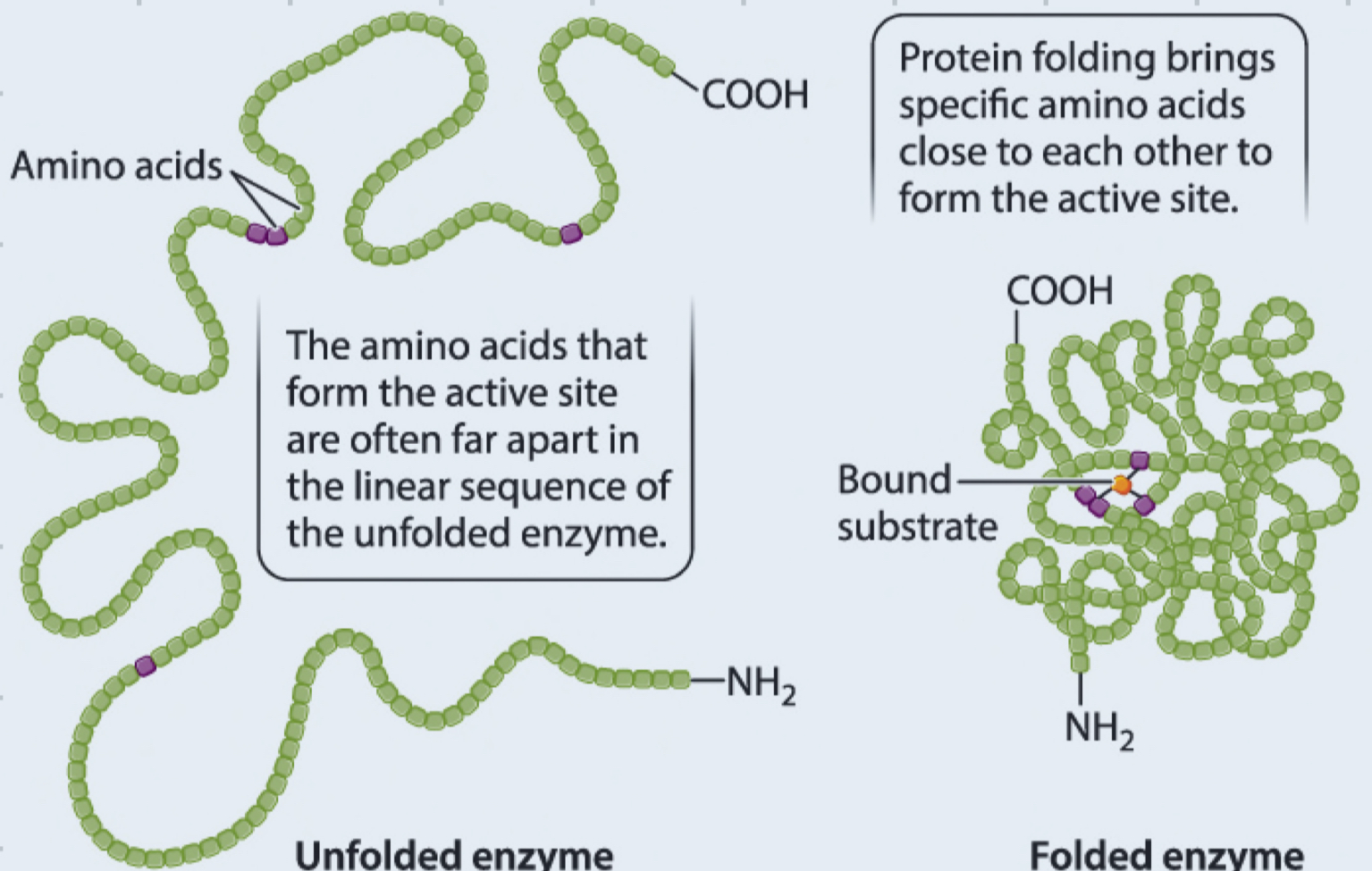
How does protein folding allow for enzyme specificity?
enzymes are proteins consisting of amino acids
protein folding brings specific amino acids close to each other to form the active site
active sites are highly specific and only bind certain substrates for certain reactions
3D structure of enzyme, determined by its AA sequence, allows it to interact with its specific substrate in a complementary manner
lock-and-key model
induced fit model
How are enzymes regulated? What are the types of inhibition? How are enzyme inhibitors used as drugs?
enzymes are regulated by activators which increase activity and inhibitors which decrease activity
competitive inhibition: inhibitor binds to active site, competing with substrate and reducing the rate of the the reaction
allosteric/non-competitive inhibition: inhibitor binds to allosteric site, changing the shape of the active site and therefore reducing the rate of the reaction
enzyme inhibitors as drug to treat alcoholism: disulfiram
disulfiram works by inhibiting the enzyme “aldehyde dehydrogenase”, causing many of the effects of a hangover to be felt immediately following alcohol consumption
likely prevents alcohol consumption
What is cellular respiration? What are the 4 major steps?
Cellular Respiration: a series of catabolic reactions (release of energy) that convert the energy stored in food molecules, like glucose, into the energy stored in ATP
Step 1: Glycolysis: occurs in cytoplasm, glucose is broken down into 2 pyruvate, producing small amount of ATP & NADH
first step requires ATP in order to charge molecules
Step 2: Pyruvate Oxidation: occurs in mitochondrial matrix, pyruvate is converted into acetyl-CoA releasing CO2 and producing NADH
Step 3: Citric Acid Cycle (Krebs Cycle): occurs in mitochondrial matrix, acetyl-CoA enters cycle, producing ATP, NADH, FADH2, and CO2
Step 4: Electron Transport Chain (ETC): occurs in inner mitochondrial membrane, electrons from NADH and FADH2 are passed along a series of protein complexes, generating a proton gradient that drives ATP synthesis
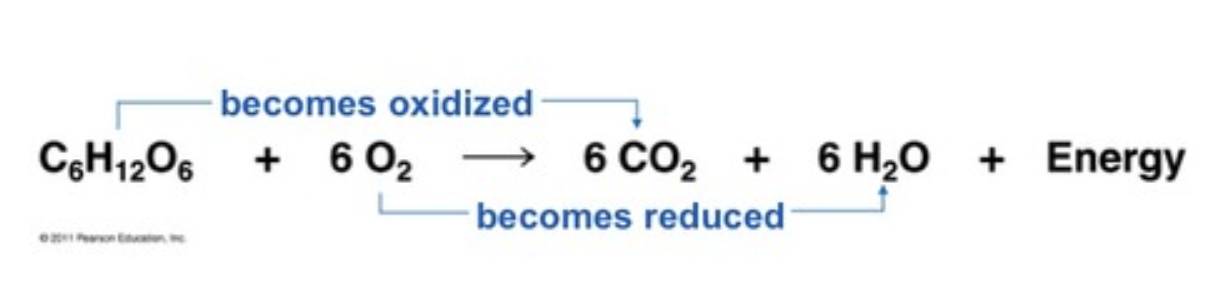
What are oxidation-reduction reactions? How is glycolysis
Oxidation: loss of electrons, often associated with the loss of hydrogen atoms
electron carriers in their oxidized form do not have H
i.e., NAD+, FAD
LEO = Loss of Electrons is Oxidation
Reduction: gain of electrons, often associated with the gain of hydrogen atoms
electron carriers in their reduced forms contain H
i.e., NADH, FADH2
GER = Gain of Electrons is Reduction
LEO the lion says GER
Glycolysis as a Redox Reaction:
glucose is oxidized to produce CO2 and H2O in cellular respiration, electrons are transferred from glucose to oxygen, releasing energy
How is ATP generated in cellular respiration? (2 different ways)
substrate-level phosphorylation: ATP is directly generated during glycolysis and the citric acid cycle
oxidative phosphorylation: ATP is indirectly generated through the flow of electrons down the electron transport, driving ATP synthase
oxygen is input into the ETC
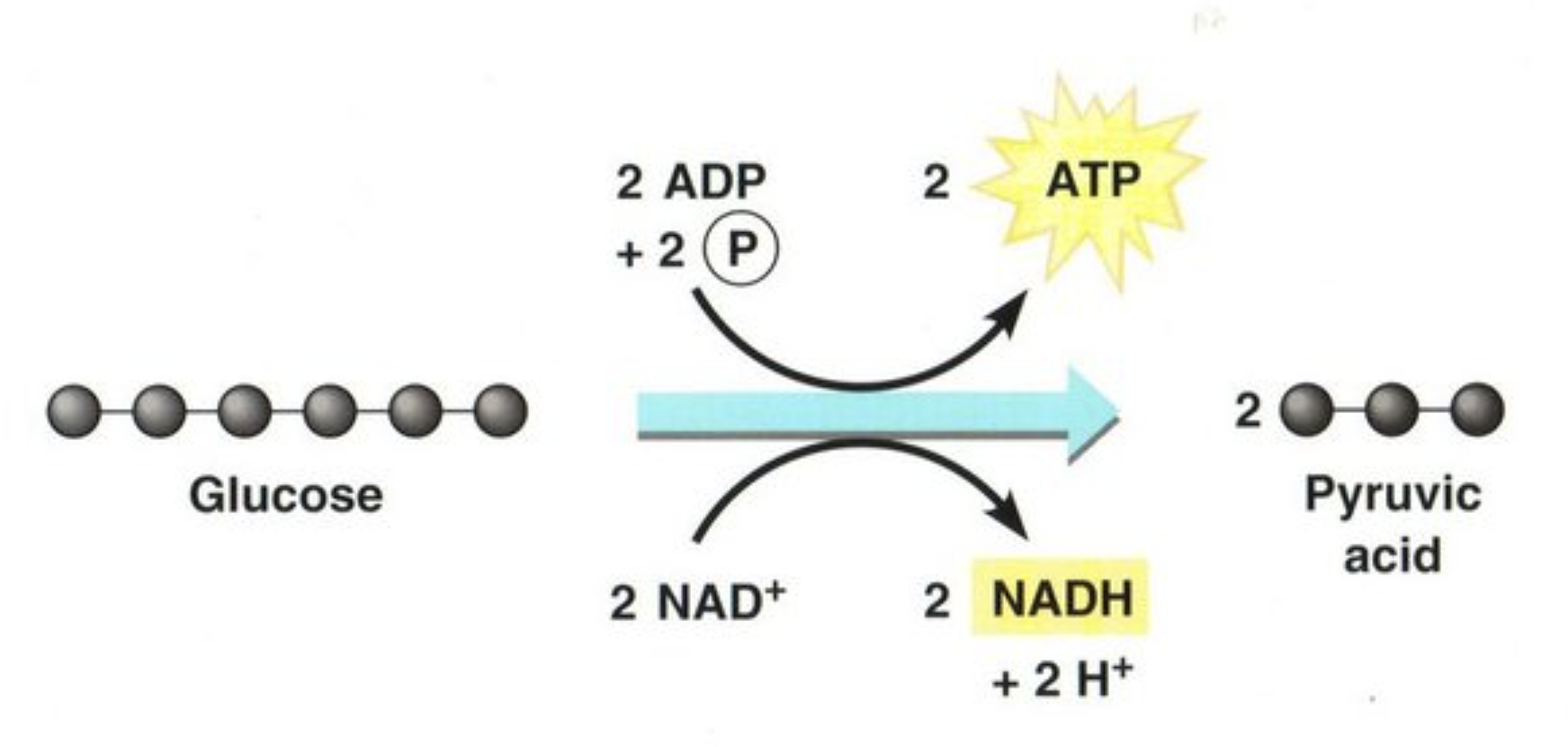
What is the chemical equation for glycolysis? What are the energy-storing molecules that are produced?
Glucose + 2 NAD+ + 2 ADP + 2 Pi —> 2 Pyruvate + 2 NADH + 2 H+ + 2 ATP + 2 H2O
During glycolysis, glucose breaks down into pyruvate and energy; a total of 2 ATP is derived in the process
2 ATP and 2 NADH energy storing molecules are produced
What are two metabolic pathways that pyruvate can enter after glycolysis?
Aerobic Respiration: pyruvate enters the citric acid cycle after being converted to acetyl-CoA
Anaerobic Respiration/Fermentation: in the absence of oxygen, pyruvate is converted to either lactate or ethanol and CO2
What are the products of the citric acid cycle?
NADH, FADH2, ATP and CO2
only FADH2 is produced in the citric acid cycle
How does the movement of electrons along the electron transport chain lead to the generation of the proton gradient?
ETC is a series of protein complexes in the inner mitochondrial membrane
electrons from NADH and FADH2 are transferred along protein complexes, releasing energy
this energy is used to pump protons protons (H+) across the membrane which creates the proton gradient
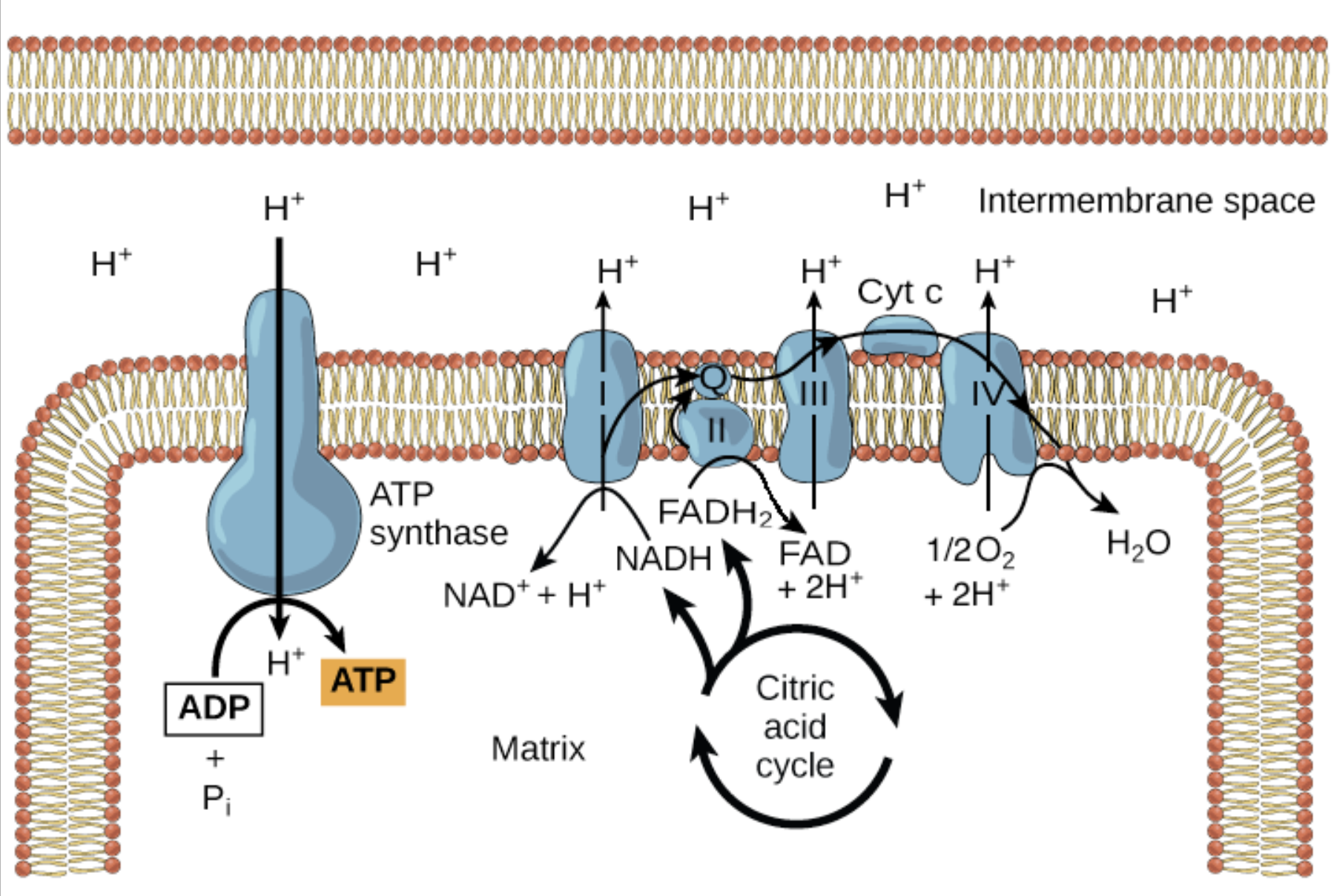
How is the proton gradient used to generate ATP?
the proton gradient drives protons back across the mitochondrial membrane through ATP synthase
ATP is generated from ADP + Pi (inorganic phosphate)
How do electrons and protons flow in the mitochondrial electron transport system
electrons move from NADH and FADH2 through Complex I and Complex II, when reaching Complex IV electrons combine with oxygen to form water
along chain, protons are pumped across membrane, contributing to the proton gradient
What are lactic acid fermentation and alcoholic fermentation?
Lactic Acid Fermentation: pyruvate is converted to lactate, regenerating NAD+ for glycolysis
i.e., occurs in muscles during exercise when oxygen is limited
Alcoholic Fermentation: pyruvate is converted to ethanol and CO2, regenerating NAD+ for glycolysis
i.e., yeast in some bacteria
What are common misconceptions relating to photosynthesis?
1. “Photosynthesis is the opposite of respiration”
this is not true. looking at their equations, they differ only in the form of energy that is being absorbed or released. however, they are not the reversal/opposite of each other as each one takes place in its own particular series of steps
they are different but not the reverse
2. “Photosynthesis is carried out in place of cellular respiration”
this is not true because photosynthesis and cellular respiration occur at the same time because photosynthesis does not generate enough ATP by itself
3. “Carbon dioxide is converted to oxygen during photosynthesis”
this is not true. just because CO2 is an input and oxygen is an output doesn’t mean that CO2 is converted into O2, there are many steps in between
4. “Plants get most of their food from the soil”
this is not true as plants get a majority of their energy from producing glucose in photosynthesis, not soil.
5. “Plants mass comes from water and minerals”
this is not true as the mass is primarily carbon which comes from CO2 used during photosynthesis
plants convert the sun's energy into chemical energy which is captured within the bonds of carbon molecules built from CO2 and water
Why does water as an electron donor require the photosynthetic ETC with the two photosystems?
water is a reliable and abundant source of electrons
a single photosystem cannot harvest enough energy
in photosystem II energy is absorbed by electrons, creating electrons which are passed on to the electron transport chain but in photosystem I the electrons are picked up by NADP to form NADPH
electrons pass through photosystems and are transferred all the way from water to NADPH
How does linear electron transport compare to cyclic electron transport?
Linear: electrons flow from Photosystem II to Photosystem I and eventually produce NADPH by reducing NADP+ in addition to ATP
Cyclic: electrons circulate within Photosystem I, generating ATP but not NADPH
What is the equation for photosynthesis? What is oxidized? What is reduced?
light energy + 6 CO2 + 6 H2O —> Glucose + 6 O2
glucose and CO2 are produced via photosynthesis
oxidized (LEO): H2O lose electrons and produce O2
reduced (GER): CO2 gain electrons to form glucose (C6H12O6)
What are the differences/similarities between photosynthesis and cellular respiration?
Photosynthesis:
converts light energy into chemical energy (glucose)
takes place in chloroplasts
consumes CO2 and produces O2
Cellular Respiration:
converts chemical energy (glucose) into ATP
takes place in mitochondria
consumes O2 and produces CO2
What are the inputs and outputs of the Calvin Cycle?
inputs: CO2, ATP, NADPH
outputs: Glucose, ADP, NADP+, Pi (inorganic phosphate)
What are the 3 main steps of the Calvin Cycle? What is the role of rubisco?
Step 1: Carboxylation: CO2 is added to RuBP (5-carbon sugar) to make a 6 carbon molecule with the help of the enzyme Rubisco
Step 2: Reduction: ATP and NADPH convert 3-PGA into G3P
Step 3: Regeneration: RuBP is regenerated for further CO2 fixation
What are antenna chlorophylls and reaction centers?
antenna chlorophylls: pigments in the photosystems light-harvesting complex — capture energy and funnel it towards reaction center
reaction centers: donate electrons to the electron transport chain