bio ap exam review
1/124
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
125 Terms
asexual reproduction
doesn’t require production or fusion of gametes. a mitosis-like process to form 2 genetically identical offspring. fast, adaptable, no mate needed, can lead to a higher population but limits resources and as no genetic diversity
sexual reproduction
produces and fuses gametes, uses both meiosis and mitosis. the parents will copulate, undergo fertilization, form single celled zygote and go through growth and differentiation, allowing for genetic diversity. takes more time and offspring is not adapted at birth
haploid gametes
a sperm or egg cell that has ½ the normal number of chromosomes (which is 23 in humans).
diploid somatic cells
a body cell with a full set of chromosomes (46)
meiosis
process by which sexually reproducing organisms produce genetically diverse haploid gametes
homologous chromosomes
two chromosomes that possess the same structure and genes (twinsies) and the cross over to exchange genes in meiosis (prophase 1 only), causing genetic diversity
steps of meiosis 1
interphase (containing G1, S (for DNA replication), and G2)
prophase 1: crossing over of Homologous chromosomes
metaphase 1: H chromosomes line up in the middle of the cell
anaphase 1:
interphase
stage of cell cycle including G1, G2, and S phase. It is the phase where the cell prepares for division by growing and replicating its DNA. 23/24 total hours in cell cycle
mitosis
the process of cell division that results in two identical daughter cells, consisting of prophase, metaphase, anaphase, and telophase. 1/24 hours in cell cycle.
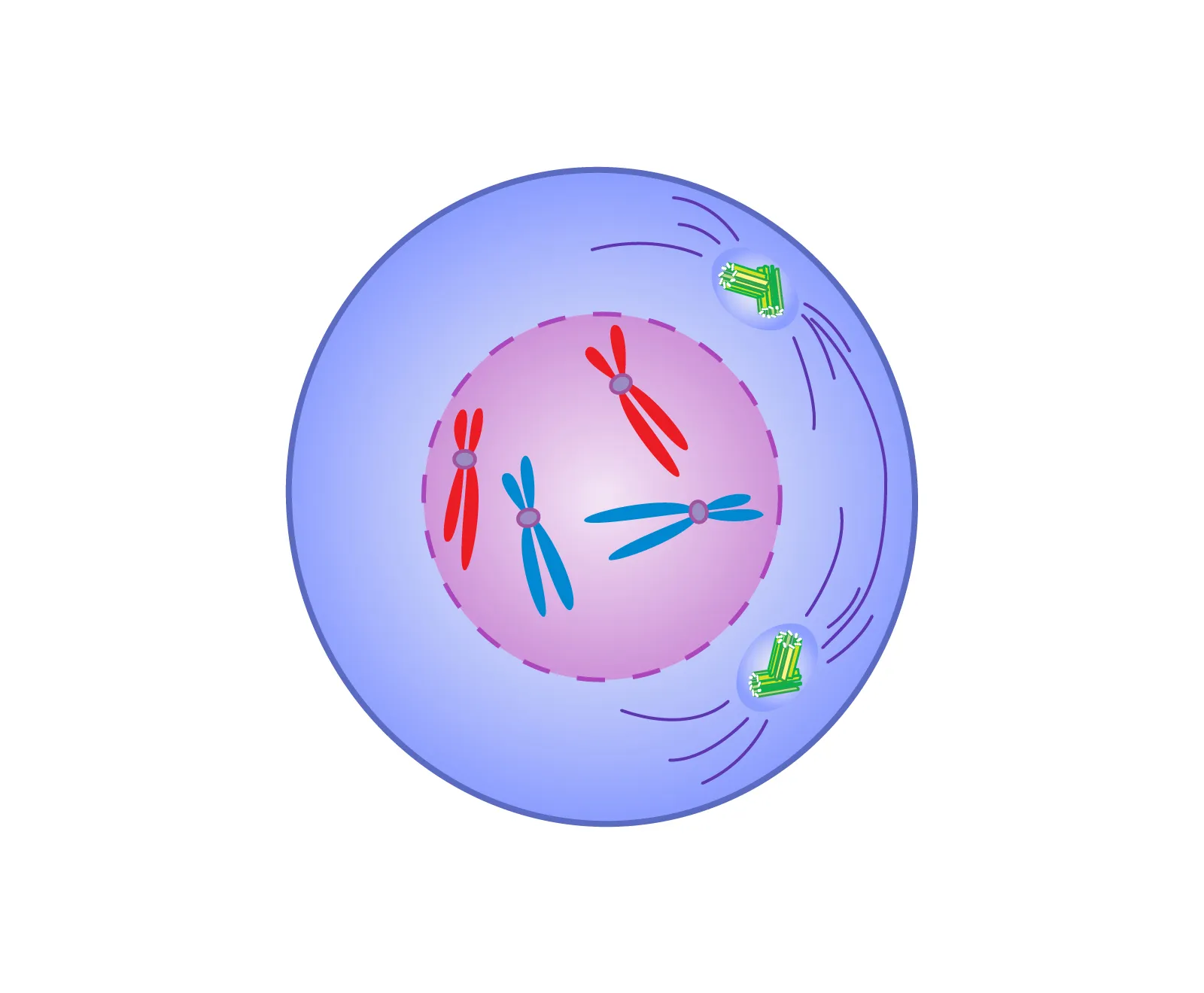
prophase
nucleus dissolves, DNA condenses into visible chromosomes and spindle fibers begin to form.
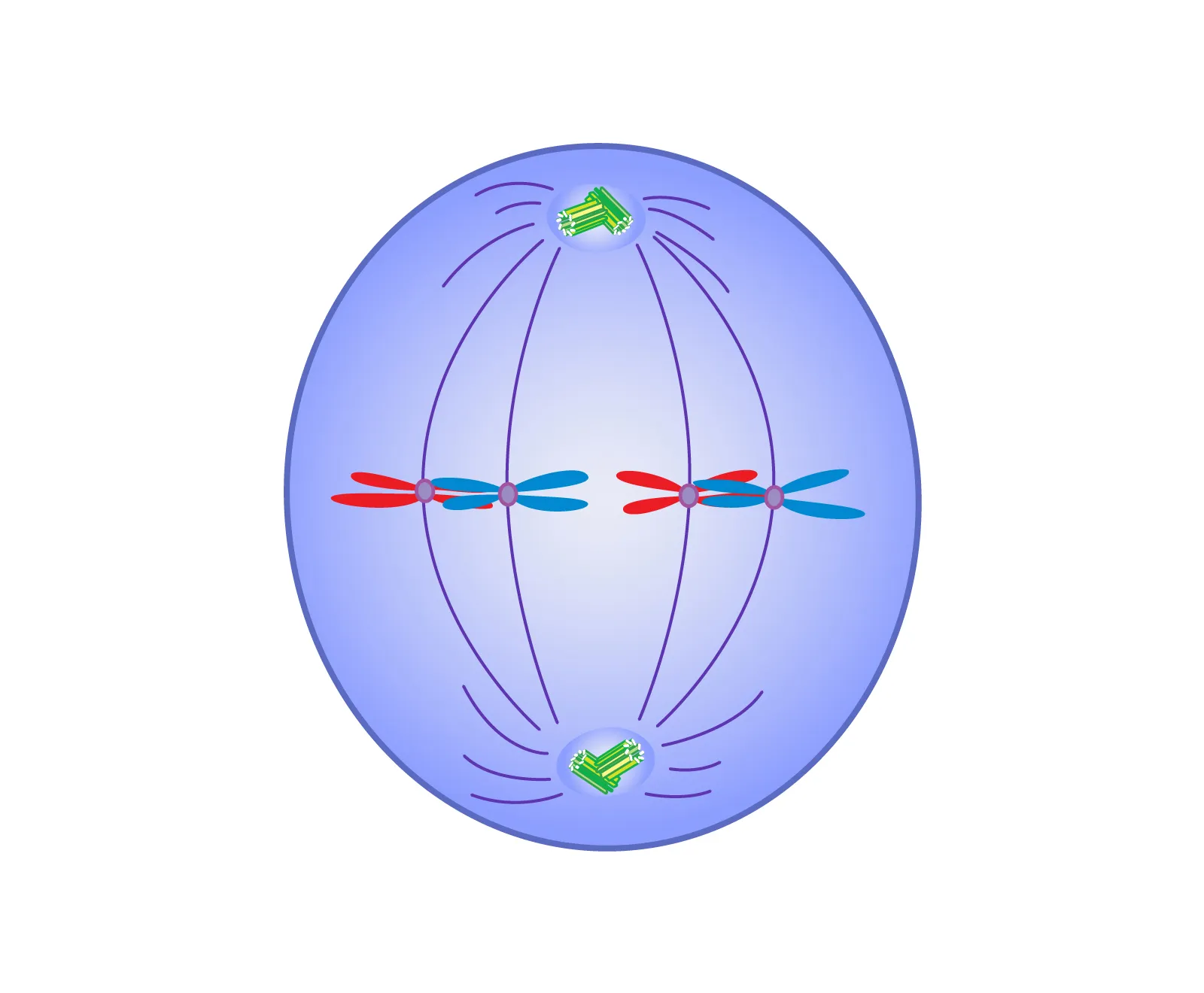
metaphase
the stage of mitosis where chromosomes align at the cell's equatorial plane, and spindle fibers attach to their centromeres.
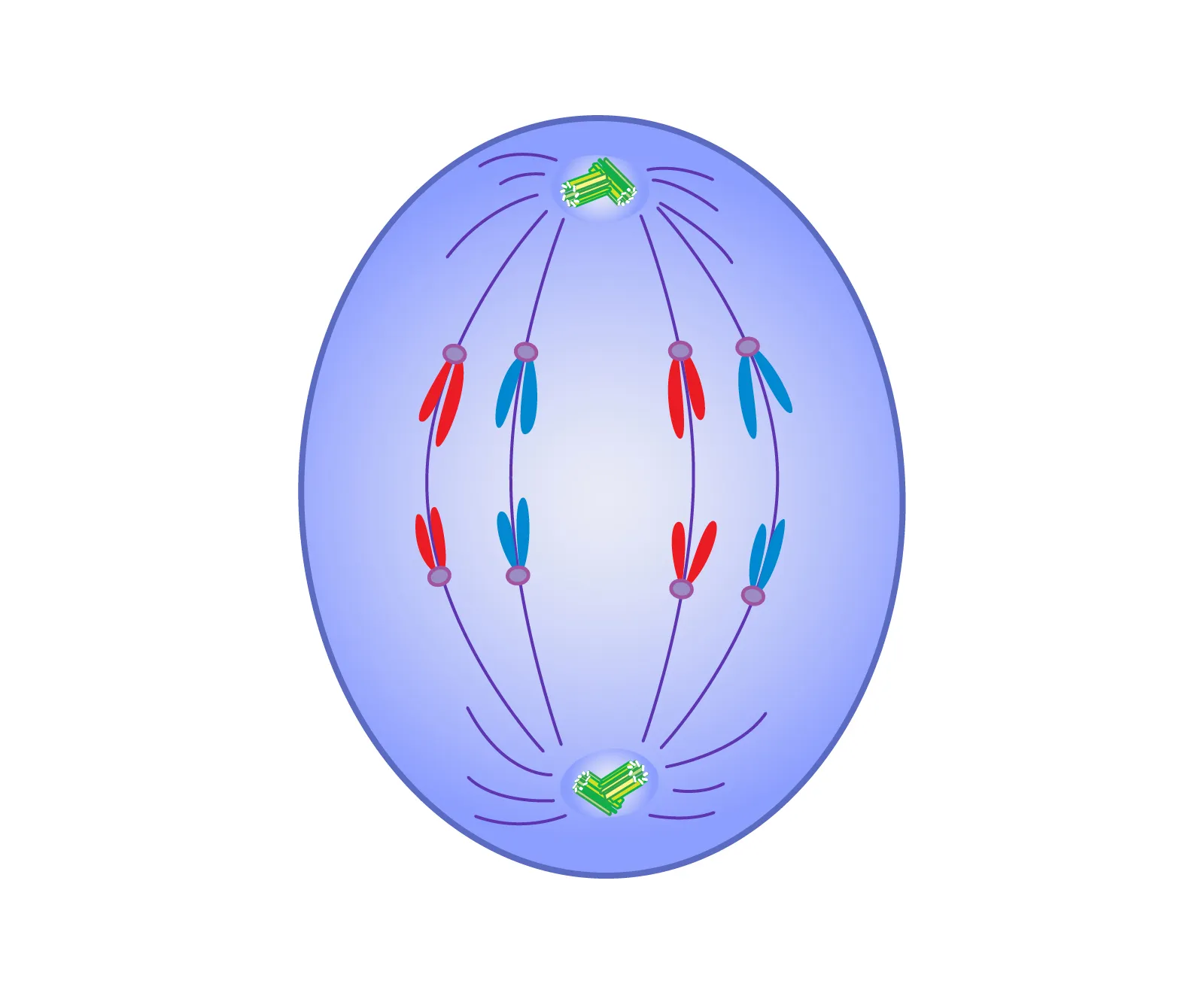
anaphase
spindle fibers contract to pull chromosomes apart at the centromere, sister chromatids moved to opposite poles of the cell, ensuring each daughter cell will receive an identical set of chromosomes.
telophase
the final stage of mitosis where the chromosomes decondense, nuclear envelopes reform around each set of chromosomes, and the cell prepares to divide.
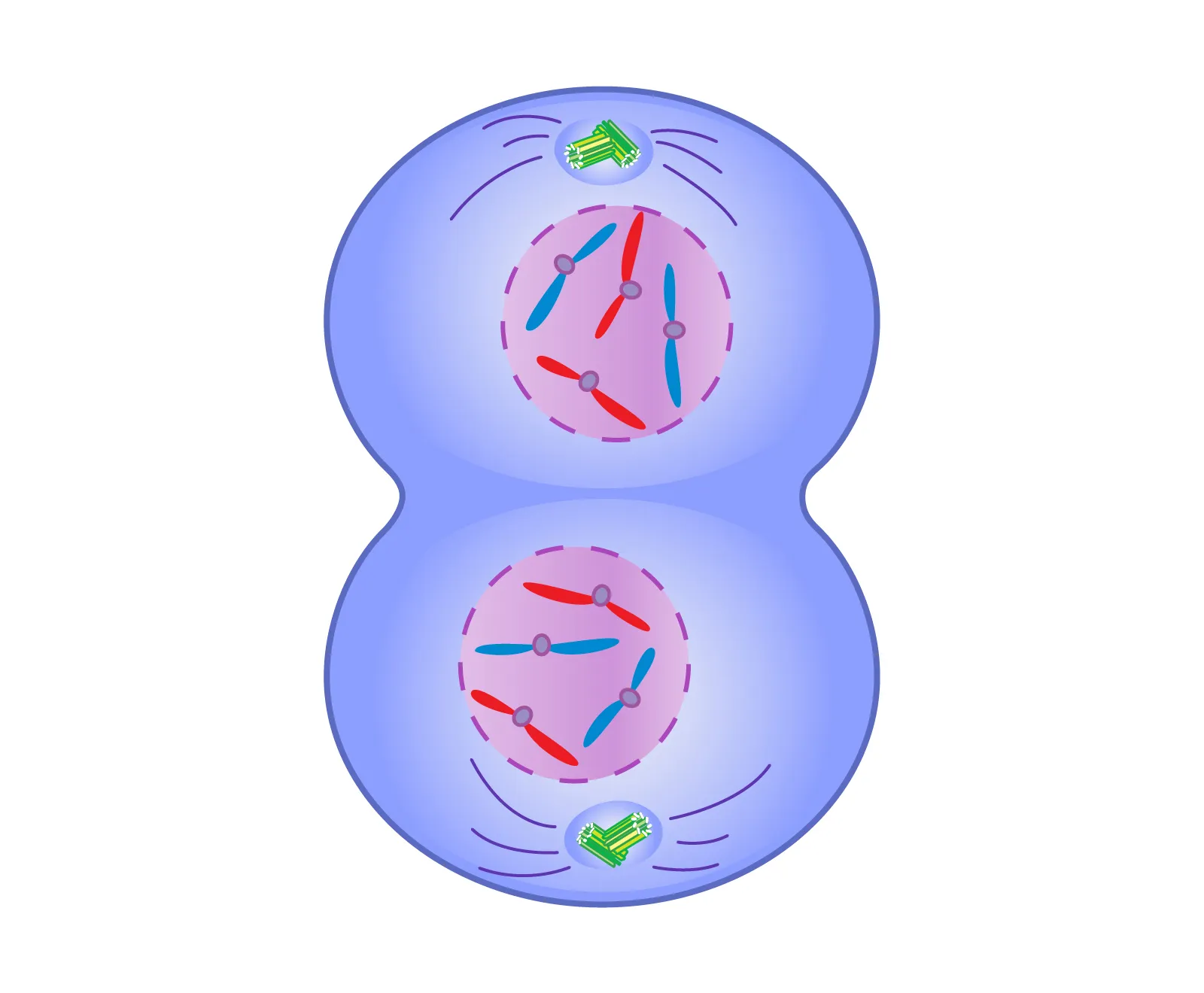
cytokinesis
the process that follows telophase, where the cytoplasm divides, resulting in two separate daughter cells, each with a complete set of organelles and genetic material.
cyclins
are regulatory proteins that control the progression of the cell cycle by activating cyclin-dependent kinases (CDKs), ensuring proper timing of cell division. specific to certain CDKs, which is important to the regulation of various phases in the cell cycle.
phosphorylation cascade
is a series of biochemical events where one enzyme activates another through the addition of a phosphate group, amplifying a cellular signal. caused by cyclin binding to CDKs and allows cell to pass through checkpoints and continue through the cell cycle.
checkpoints of cell cycle
are regulatory points in the cell cycle that ensure proper division by checking for DNA damage, proper chromosome alignment, and mutations before allowing the cell to proceed to the next phase.
p53
is a tumor suppressor protein that plays a critical role in the cell cycle by preventing the proliferation of cells with damaged DNA. It activates DNA repair proteins, induces cell cycle arrest, and can trigger apoptosis if damage is irreparable.
sister chromatid
is one of the two identical halves of a duplicated chromosome that are joined together at the centromere, separated during cell division (1/2 of chromosomes)
eukaryotic DNA
is organized into linear chromosomes within a nucleus, and is associated with histone proteins that help in packaging and regulating gene expression.
prokeryotic DNA
is organized in a circular form and is not enclosed within a nucleus. It typically exists as a single, double-stranded DNA molecule and is found in prokaryotic organisms such as bacteria.
DNA nucleotides’ chemical groups
includes a phosphate group attached to 5’ deoxyribose, a 5-carbon Deoxyribose sugar (numbered for identification), and 1 of 4 nitrogenous bases.
nucleotides pairing
A forms 2 H bonds with T (thymine), and C forms 3 H bonds with G (guanine). G and A are purines (double ring), while C and T are pyrimidines (single ring).
Chargaff’s rule
states that in any given DNA molecule, the amount of adenine ~ thymine, the amount of cytosine ~ guanine, the amount of purine = pyrimidine, and A+T doesn’t = C+G
telomeres
are repetitive DNA sequences at the ends of eukaryotic chromosomes that protect them from deterioration or fusion with neighboring chromosomes.
DNA replication stages
1) initiation: topisomerase relaxes the supercooled DNA and unwinds it, breaking the Hydrogen bonds between template strands and single stranded binding proteins keep it from reannealing; 2) elongation: primase puts down RNA primers to help DNA polymerase bind. DNA polymerase then synthesizes new strands in a 5’-3’ direction; 3) termination: RNA primers are removed and ligase fills in gaps from primer removal and between okazaki fragments. the end result is 2 identical copies and reformation of the double helix.
feedback mechanisms
sensory pathways/responses your body uses to maintain homeostasis
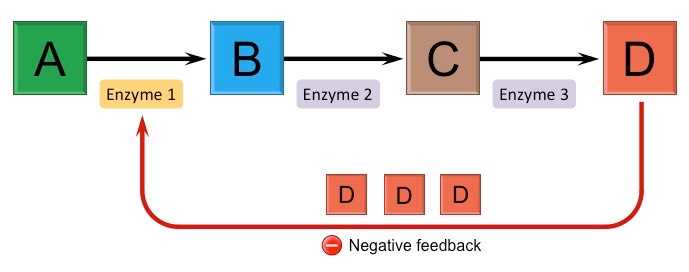
negative feedback mechanism
a process that counteracts a change in a physiological variable, helping to maintain homeostasis by reversing the direction of change — decreases a response.
positive feedback mechanism
a process that amplifies a change in a physiological variable, leading to an increase in the response and moving the system away from its starting state.
ligand
signal molecule that binds to a receptor to trigger a response in a cell.
short range signaling
a communication method where signaling molecules affect nearby cells, often through diffusion, to elicit a quick response.
paracrine signaling
short range signaling where cells communicate with nearby cells through the release of signaling molecules.
autocrine signaling
short range signaling where a cell secretes signaling molecules that bind to receptors on its own surface, influencing its own activity. the target cells and signal cell are the same/similar
long range signaling
a type of cell signaling that sends signals to different areas of the body
cell-cell contact
signaling with direct contact of signal proteins to receptor proteins
no cell-cell contact
a ligand is released and travels to the target cell through blood stream
gap junction signaling
in animal cells, where cell membranes fuse and pores open to send signals with cell-cell contact
plasmodesmata
in plant cells, cell walls and membranes fuse to signal with cell-cell contact.
endocrine signaling
hormones are sent out into the bloodstream and ligands travel throughout the body to send signals through no long range signaling
neuronal signaling/synaptic signaling
neuron’s axonal projection transmits signals throughout the body through long range signaling
quorum sensing
allows bacteria to communicate with each other and respond to changes in cell population density be regulating gene expression
receptor
intracellular or membrane bound proteins that bind to ligands and undergo shape changes to transmit signal inside a cell
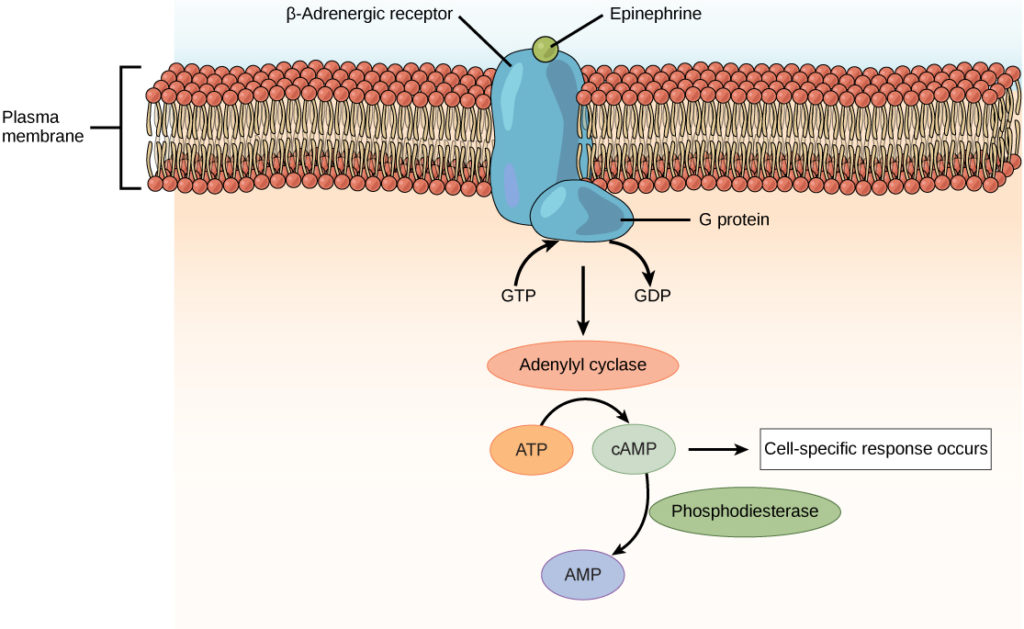
membrane bound receptors
hydrophilic ligands bind to this kind of receptor embedded in the cell membrane (ligands does not cross the membrane). the signal is amplified and moves into the cell via a transduction pathway
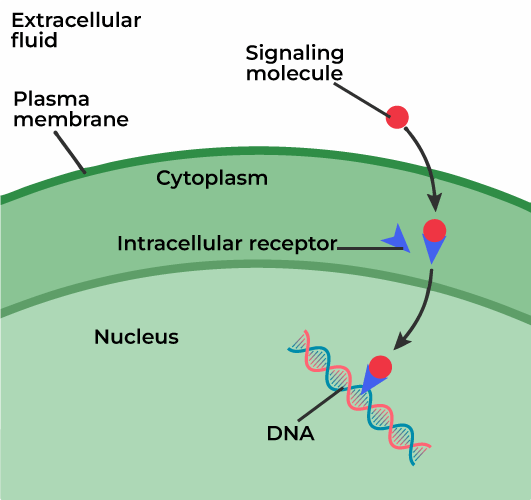
intracellular receptors
lipid soluble (hydrophobic) ligands bind to this kind of protein in the cytoplasm or nucleus of a cell by diffusing into the cell. if in cytoplasm, forms a complex that travels to the nucleus and bypasses transduction. steroid and thyroid hormones can easily by diffused into cells this way. an activated hormone-receptor complex can turn on or off specific genes (transcription factor).
ion channel linked
membrane bound receptor that directly opens or closes ion channels in the cell membrane when a ligand binds, which allows ions to flow across the membrane
g-protein coupled
membrane bound receptor that interacts with g-proteins in the membrane (coupled) to signal within cells.
enzyme-linked receptor
membrane bound receptor that has intrinsic enzymatic activity and activates when a ligands binds. initiates phosphorylation cascades and other signaling events
transduction pathway
moves signal from the cell membrane to the nucleus to amplify the signal and sometimes produce second messengers.
phosphorylation cascade
adding and removing phosphates, regulate protein activity. most relay molecules in these pathways are protein kinases. turn on/off molecular activities
second messengers
small, nonprotein, water soluble molecules or ions that spread through a cell via diffusion. participate in pathways initiated by other molecules like RTKs. remember Cyclic AMP, Cyclic GMP, and calcium ions.
molecular responses to cell signaling
changes in gene transcription or enzyme activity
Receptor Tyrosine Kinase pathway
a network of transmembrane proteins that regulate cell functions like growth, metabolism, and motility. when a ligase binds to an RTK, the protein kinase in the receptor is activated and enzymatic reactions are triggered
IP3 pathway
a series of events that releases calcium Ca+ ions from the endoplasmic reticulum and the subsequent activation of various cell responses. important for drug resistance, signal translation, and more.
signaling pathway symbols
negative feedback pathways will be shown by an arrow with a flat head. two intersecting lines like this will cancel out and actually show amplification of a response. positive feedback pathways will be shown by an arrow.
cell
group of organelles and molecules working together to perform a specific task and help organism maintain homeostasis. range from 10mm to 1 mm to be big enough to fit all DNA and organelles but small enough to exchange nutrients, etc. to maintain homeostasis.
prokaryote
bacteria and Archaean, singe celled. no nucleus, no membrane bound organelles. oldest type of cell. some have cell walls and flagella. contains DNA/RNA, has cell membrane, ribosomes, and uses cellular respiration.
eukaryote
newest type of cell that makes up all plants, animals, fungi, and protists. can be single or multicellular. has nucleus, cell walls, flagella, DNA/RNA, cell membrane, and membrane bound organelles.
non membrane bound organelles
ribosomes, cell membrane, cell wall, cytoplasm, cytoskeleton, centriole, spindles, flagella
membrane bound organelles
nucleus, golgi body, chloroplast, endoplasmic reticulums, vacuoles, mitochondria
compartmentalization
membrane bound organelles in cells allow for different areas of the cell to different things, meaning they can specialize in specific areas. more specialization = more complex life forms. i.e. enzymes and substrates can be located in the same place of the cell for more efficient reactions
membrane folding
organelle membranes are usually folded to increase surface area which increases the organelles ability to complete important functions/reactions without making the cell too big (volume).
endosymbiotic theory
eukaryotic cells evolved after a large ancestral prokaryote ingested mitochondrion and chloroplast like proto-prokaryotes and formed a close mutual relationship with them. the ancestral prokaryote became able to survive in the increasingly oxygen rich atmosphere, and proto prokaryotes got resources/protection from harsh conditions of early earth’s atmosphere
evidence of endosymbiosis
MADDR—membranes (double), antibiotics (cells susceptible to some), division (reproduction occurs in fission-like process), DNA (own DNA, circular, naked), ribosomes (identical to size of prokaryotic ribosomes)
cell wall
provides structure to cells and acts as permeability barrier for some substances to the internal environment. rigid, made of complex carbs, found in plants, fungi, and bacteria.
ribosomes
atom
smallest stable unit of matter that has the characteristics of its specific element. contains a nucleus with electron orbitals. protons (positive charge) and neutrons (neutral) in the nucleus, electrons (negative) in the orbitals. usually 1 ___ is neutral because the electrons = protons.
orbitals
each has a different amount of energy associated with it. closer to the nucleus = lower energy level, while farther away = higher energy level. outermost orbital contains the valence electrons. most elements want 8 electrons because this makes the stable
cations
non neutral atom: positive charge, has more electrons than protons
anions
non neutral atom: negative charge, has more protons than electrons
main elements of life
carbon hydrogen oxygen nitrogen
trace elements
elements that are found in trace amounts throughout life — calcium, sodium
carbon
basis of all life. in its inorganic form, it has to be fixed from the atmosphere by plants in photosynthesis. its then incorporated into carbohydrates, the main source of biomass. used to produce every biomolecule, recycled through decomposition. has 4 valence electrons that can be used for forming stable, covalent bonds with other molecules (group 14 on ptable). always forms 4 covalent bonds with other atoms.
nitrogen
in its inorganic form, fixed from atmosphere by bacteria and other decomposers so plants can use it. used by organisms to produce proteins and nucleic acids, recycled by decomposers. found mostly in the atmosphere
phosphorous
used to build nucleic acids and phospholipids and to make cell membranes
electronegativity
measurement of how strongly atoms attract bonding electrons to themselves (i.e how much they’ll pull electrons to themselves. determined by how many electrons are in a valence shell — the closer to 8, the more electronegative the element is.
on periodic table, electronegativity increases to the right because protons increase, and decreases down because of the larger electron cloud
electronegative elements: fluorine, oxygen, nitrogen. fluorine most, nitrogen least.
electropositivity
measurement of the ability of elements to donate (give up) electrons and form cations. these elements usually have 1-2 electrons in their valence shells and are not very electronegative.
covalent bonds
tend to form around elements in the middle of the periodic table like carbon. occur when 2 atoms share electrons. energy is stored in the bond and released when broken, helping form ATP
ionic bonds
the transfer of valence electrons from a metal to a nonmetal. weak in a solution but strong otherwise, and can dissociate in water. less common to biology
polarity
happens when a very electronegative element bonds to a more electropositive/less electronegative element. occurs when there is unequal sharing of electrons across shared covalent bonds. the partial charges on the poles attract to opposite charges on other molecules, which allows H2O to form hydrogen bonds with other H2O molecules.
a molecule can have a polar bond and not be polar because polarity requires asymmetry and a dipole moment.
hydrogen bonds
weak attraction between H bonded to an O, N or F element (electronegative) and another O, N or F atom. the H bonded to these elements will have a partial positive charge and the O, N, and F will have a partial negative charge — opposites attract. doesn’t need a lot of energy to break one, but does require a lot to break multiple (Steam engines)
structure’s importance
how atoms bond determine the overall shape of the molecule. the structure, shape, and chemical properties of a molecule determine the function of that molecule. so, the more possible shapes, the more possible functions and therefore more ability to form complex life forms
law of conservation
1) energy is always conserved in a chemical reaction
2: the amount and type of atoms are conserved in reactions
3) amount of bonds in chemical reaction are conserved
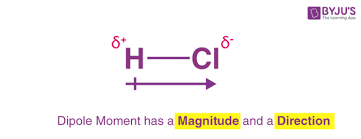
dipole moment
separation of charge around a molecule into a positive area and a negative area. the arrow in the diagram will always point to the negative side. occurs in polar molecules
ionic solid
instead of a partial + and partial -,the electrons in these solids’ molecules completely transfer into one + and one - (2 separately charged atoms) creating two charged ions like Na+ and Cl-
hybrid molecules
polar and nonpolar substance like dish soap
Van Der Waals
weak force between 2 or more nonpolar molecules. the temporary attraction of one molecule’s electron rich region and another’s electron poor region creates this intermolecular force between molecules
carbohydrates
sugars— monomers are monosaccharides (glucose), polymers are polysaccharides (complex carbs like starches, cellulose).
Hexamer ring structure — C, H, O
1C:2H:1O ratio
purpose: short term energy, energy storage, structure.
energy storing carbs have a branched structure — more branches means more monomers can be broken off, allowing for cellular respiration to happen faster and make more ATP
structural carbs have a linear structure that can stack, giving the molecules stability so they can form tough structures for support (plant cell walls)
lipids
fats/oils — monomers are fatty acids, polymers are lipids. phospholipids make up main part of cell membrane. hydrophobic.
structure is long hydrocarbon chains. C, H, O, sometimes P
1C:2H:<O ratio
saturated fat: bad fat, no double bond in molecular structure = linear, stack at room temperature so they can build up in blood vessels
unsaturated fat: have double bonds, prevents stacking, liquid at room temperature, healthier
proteins
meats/muscles — monomers are amino acids, polymers are polypeptides (proteins). amino acids like to form polypeptide chains, joined via condensation reactions with peptide bonds.The main function is wound/tissue repair. catalyzes chemical reactions, cell signaling.
complex, 4-leveled structure — C, H, O, N, S. 4 regions around a central carbon → amino group, carboxyl group, hydrogen group, variable R group
r group can be any of 20 unique side chains categorized by their chemical properties — hydrophilic (polar), hydrophobic (nonpolar), charged (ionic). interaction of the r groups affect the tertiary structure of proteins.
denaturation: changes in protein shapes that can impact protein function
enzymes
R specialized proteins that speed up (catalyze) chem reactions in cells 2 maintain homeostasis. some proteins embedded in cell membrane help with transport of materials into and out of cells
nucleic acids
commonly referred to as genetic material. monomers = nucleotides. polymers are nucleic acids.
structure is CHONP
nucleotides: made up of sugar, phosphate, and a nitrogenous base linked with covalent bonds. purine, cytosine, uracil, thymine, adenine, guanine. the nitrogenous base can be a pyrimidine (CUT, double) or purine (adenine, guanine, single).
nucleic acids: DNA and RNA, both store genetic material. have directionality that impacts how DNA reparation and transcription occurs.
DNA structure
the eukaryotes (in nucleus) that make up this polymer are linear, double stranded double helix structure. the prokaryotes (in cytoplasm) are circular, double stranded, double helix. this type stores genetic code and is made up of 4 nucleotides — adenine, thymine, cytosine and guanine with deoxyribose sugar. stable.
RNA structure
single stranded. 3 kinds are mRNA, tRNA, and RRNA. all three are used for protein synthesis and are made up of 4 nucleotides — adenine, uracil, cytosine, guanine w a ribose sugar. less stable than DNA, made in the nucleus and transported to the cytoplasm.
biomolecules
organic, carbon based macromolecules that can be used and converted by living organisms. all organisms need these to survive. Carbs, Lipids, Proteins, Nucleic Acids
monomer
individual sub unit of a biomolecule
dimer
2 monomers covalently bonded together
polymer
many monomers covalently bonded together
biomolecular metabolism
combination of chemical reactions that synthesize or hydrolyze biomolecules for energy storage and release within an organism.
catabolism
a metabolic process that releases energy in the form of heat and chemical energy (ATP/ADP) so that metabolic processes can occur. it breaks biopolymers down into monomers to release energy in the form of ATP.