IMED1002 - Genomes (L27)
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
23 Terms
Genome
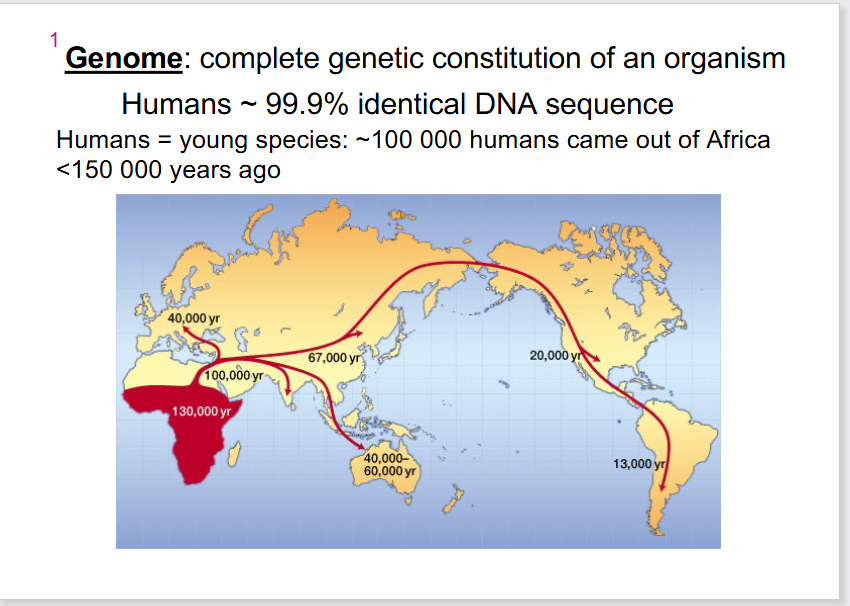
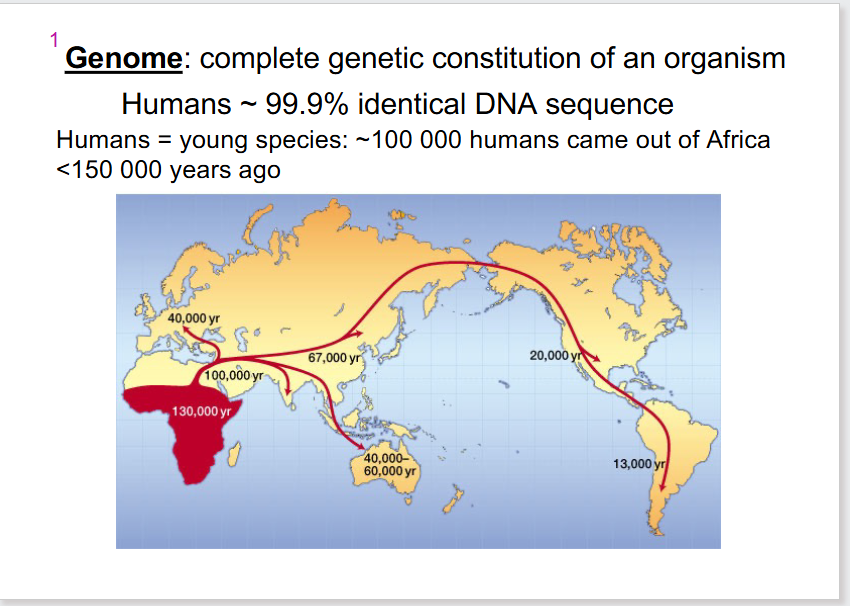
- complete genetic constitution of an organism
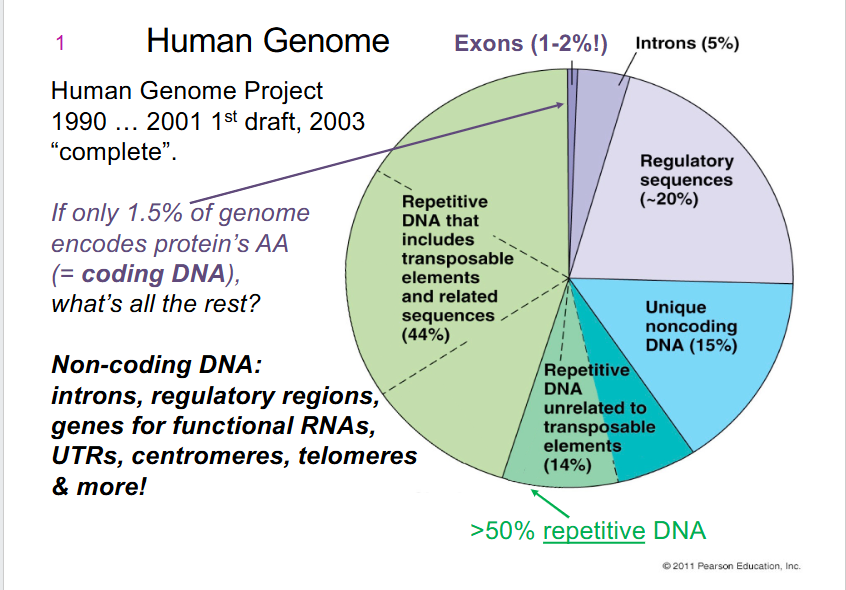
Human Genome
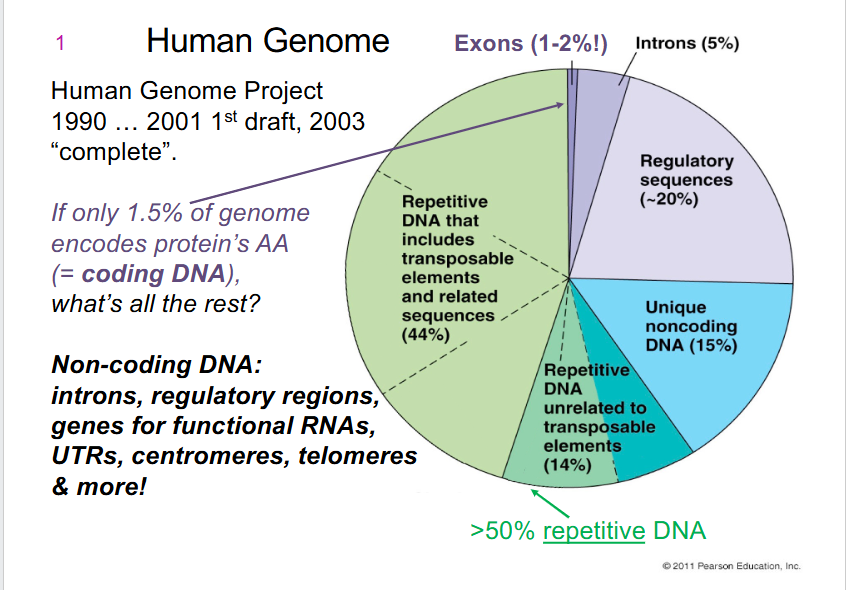
- only 1.5% of genome encodes protein's AA (= coding DNA)
- Non-coding DNA: introns, regulatory regions, genes for functional RNAs, UTRs (untranslated regions), centromeres, telomeres, more
Genome Paradox (NO LEARNING OUTCOME)
Genome size varies significantly across species, number of nt does not correlate with organ's complexity. chr number doesn't correlate either
- hence non-coding DNA must be important and essential
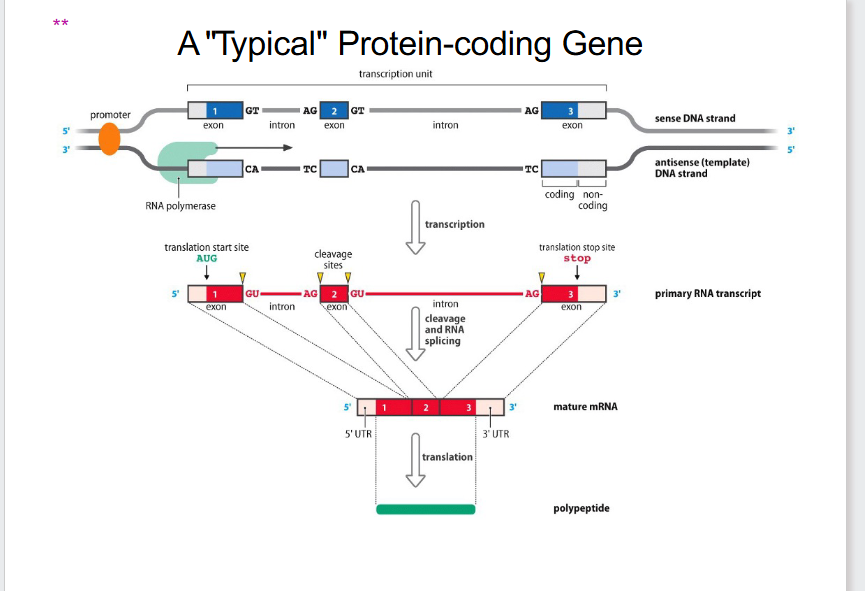
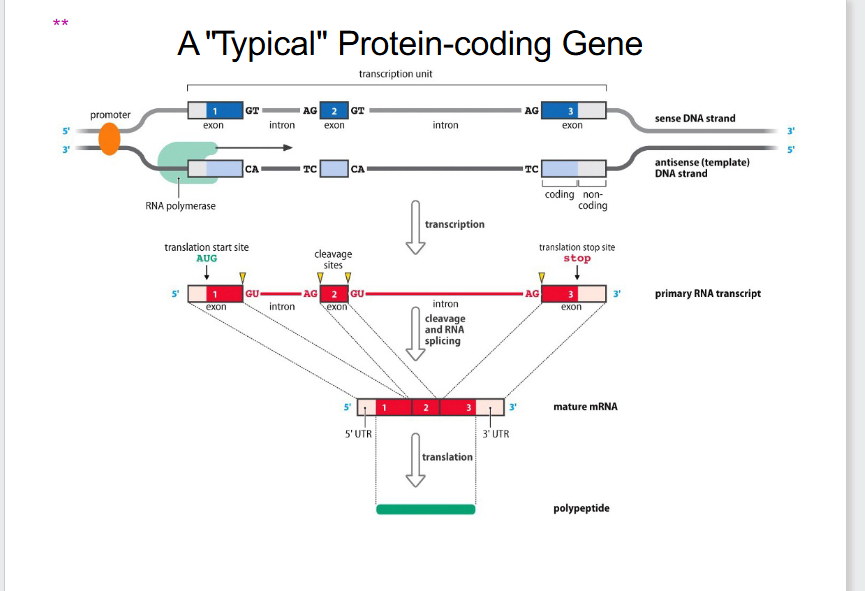
Typical Protein-Coding Gene (OLD PERSPECTIVE)
DIAGRAM ON SLIDE 8
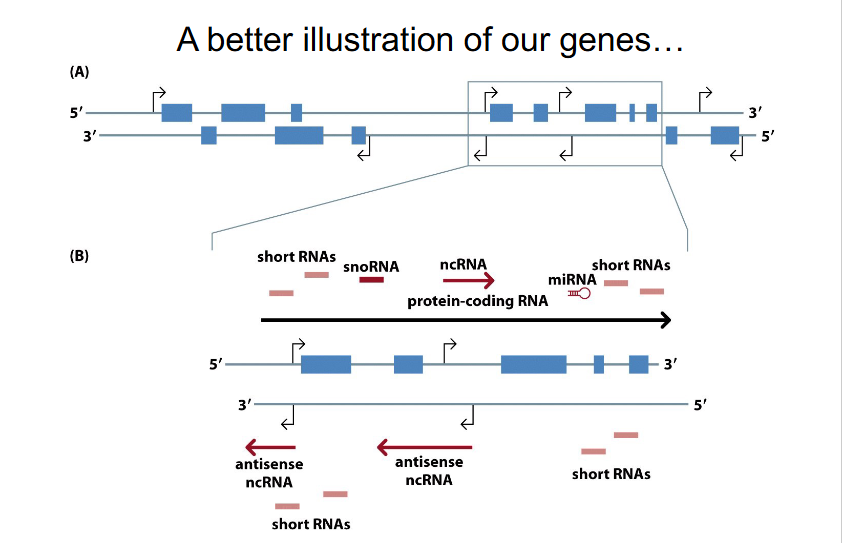
Better illustration of our genes (NEW PERSPECTIVE)
- No learning outcome
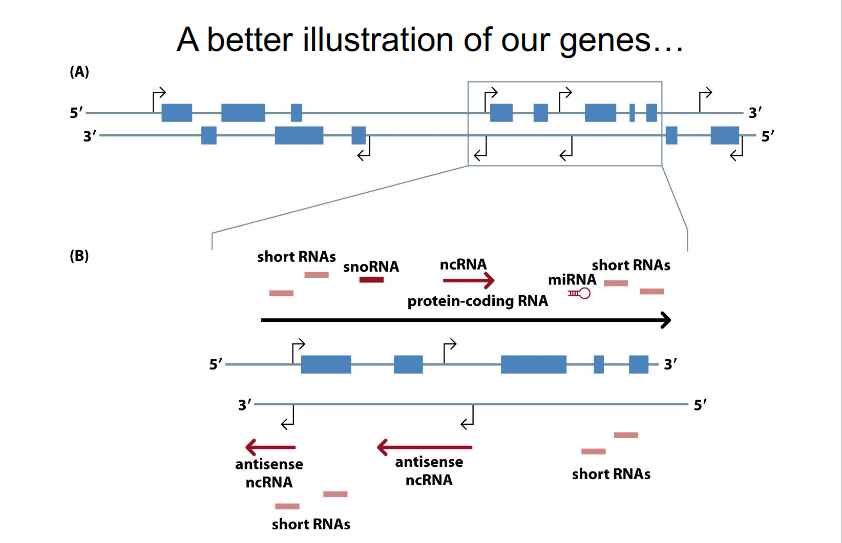
Non-coding DNA include
- introns
- regulatory regions (including promoters)
- UTRs (untranslated regions)
- genes for functional RNAs
- centromeres
- telomeres
- however, for many years the focus was on coding DNA (protein-coding genes)
Protein-coding genes
- Estimated number of keeps changing
- Before human genome sequenced (1990), estimated 45000
- in year 2000: 33,000
- Data from 2012-2018: 21306 protein coding genes, coding exons is around 1-2% of gene
- So human genome has around 20000 protein coding genes
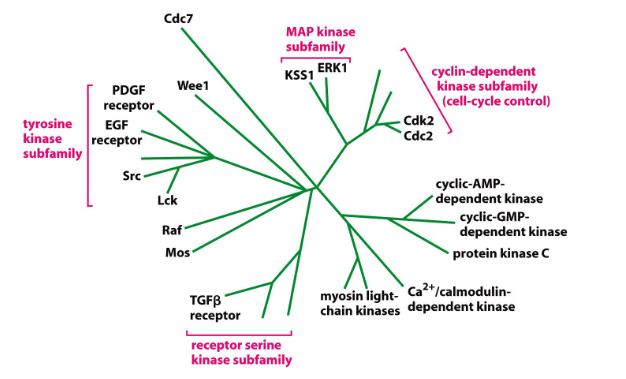
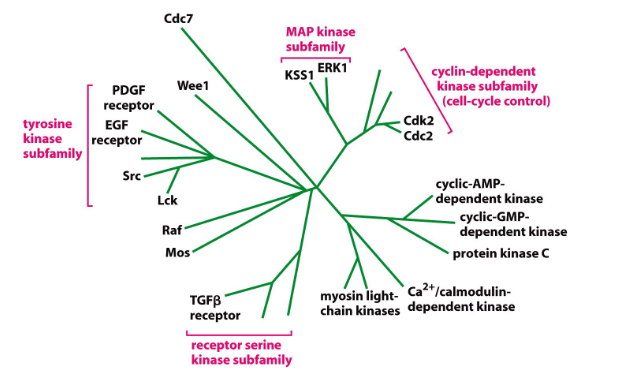
Many Proteins Belong to Families
- relationships determined by comparing AA sequences -> more similar sequences cluster on same branches of evolutionary tree. Family members share similar 3D shape and function
- based on homologous sequences
Homology
- most fundemental relationship between two proteins is homology: AA sequence similarity (based on gene's nt sequence)
Homolog
- gene/protein related to another gene/protein by descent from a common ancestral DNA sequence
- knowledge of homology helps with evolution and function
two types of homolog, based on how evolved:
- Paralog
- Orholog
Paralog
genes present within a species, derived from gene duplication
- basically they are genes that are related to one another because they evolved from gene duplication and there will be some variation between the sequences as the organism continues to evolve
Ortholog
genes found in different species derived from a single common ancestor
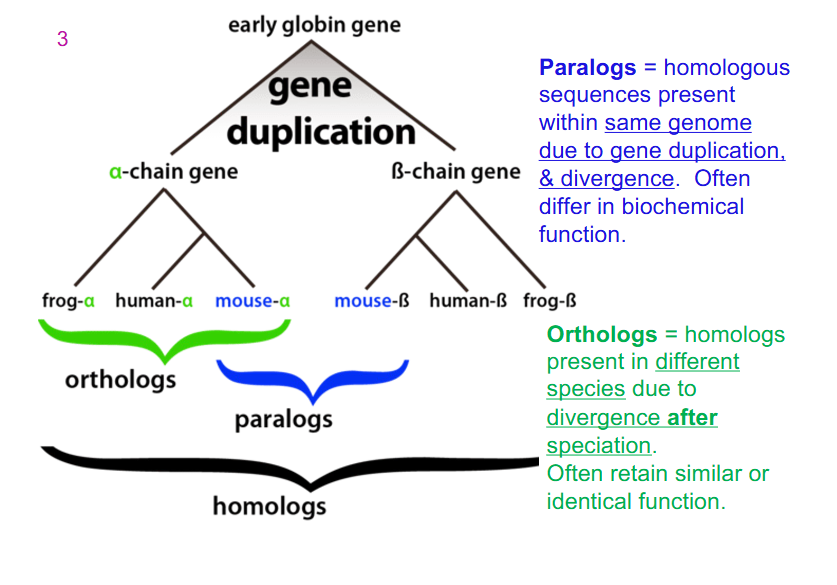
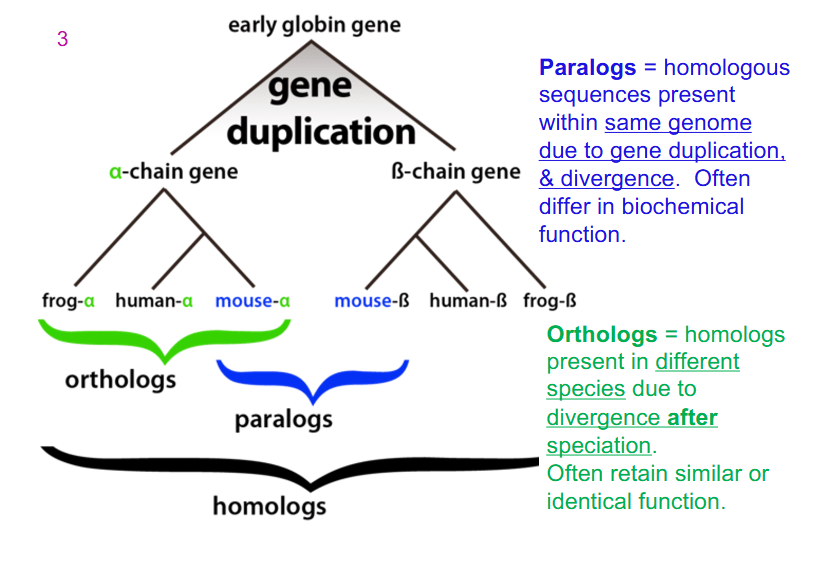
Paralogs and Ortholog in example (Early Globin Gene)
- all genes within globins are homologs: they're all derived from an early ancestor and they've undergone some changes, some variation in gene sequence over evolution
- Paralog: homologous sequences present within same genome due to gene duplication and divergence. Often differ in biochemical function
- Ortholog: homologs present in different species due to divergence after speciation. Often retain similar or identical function
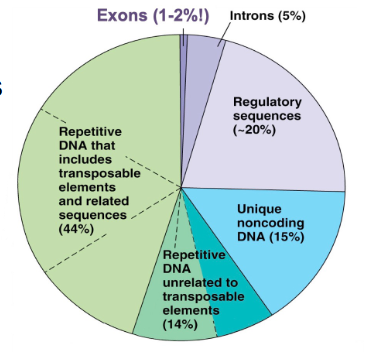
Non-coding DNA is most of our genome
- regulatory regions including promoters
- UTRs
- introns
- genes for functional RNAs
- centromeres
- telomeres
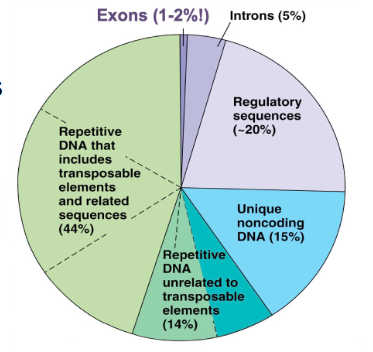
Other non-coding DNA (NAMING ONLY)
- Pseudogenes
- Repetitive Sequences
- Transposable Elements
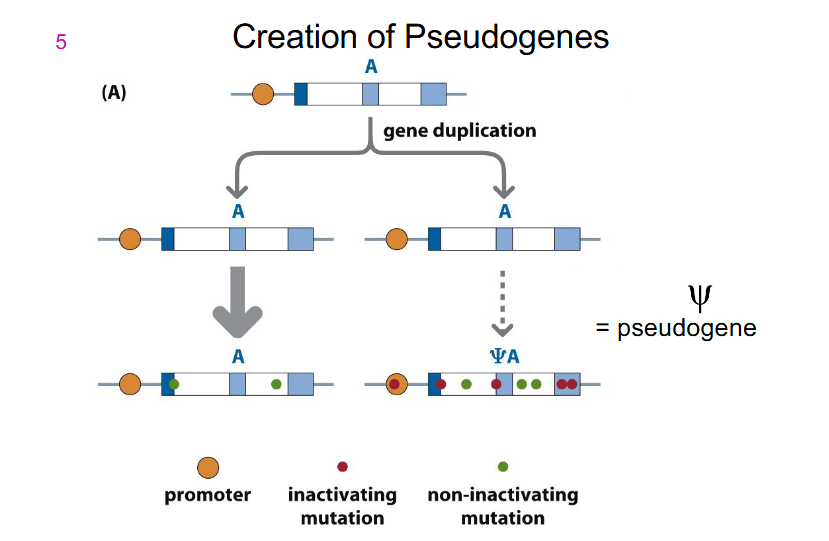
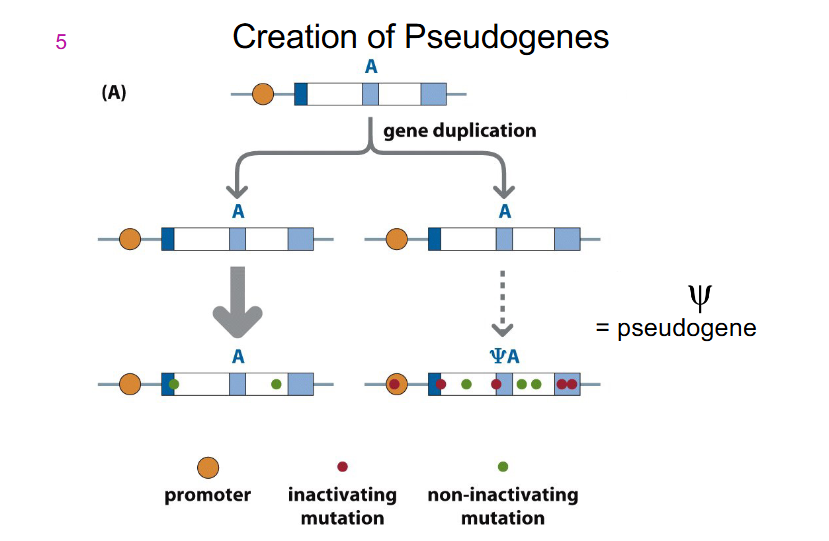
Pseudogenes
- result from duplication of genes with normal function, but the pseudogene accumulates deleterious variants and does not retain normal function
- Pseudogenes can exist because a single functioning copy of a gene is sufficient for normal cell function. Humans have around 10000 pseudogenes, some are transcribed
- designated with symbol psi
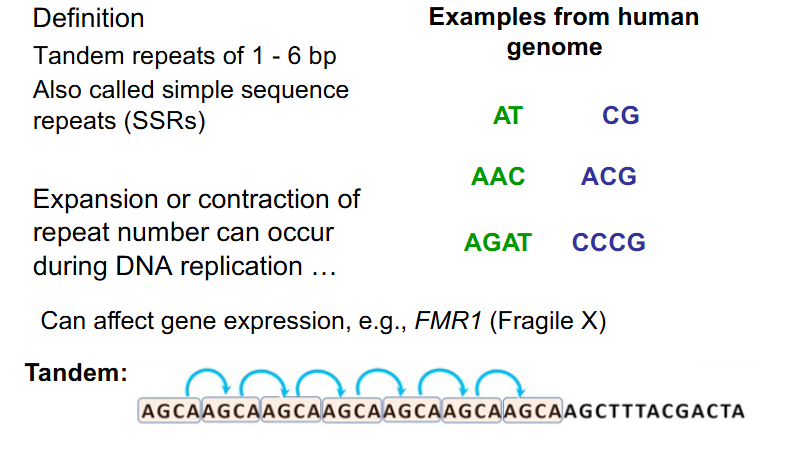
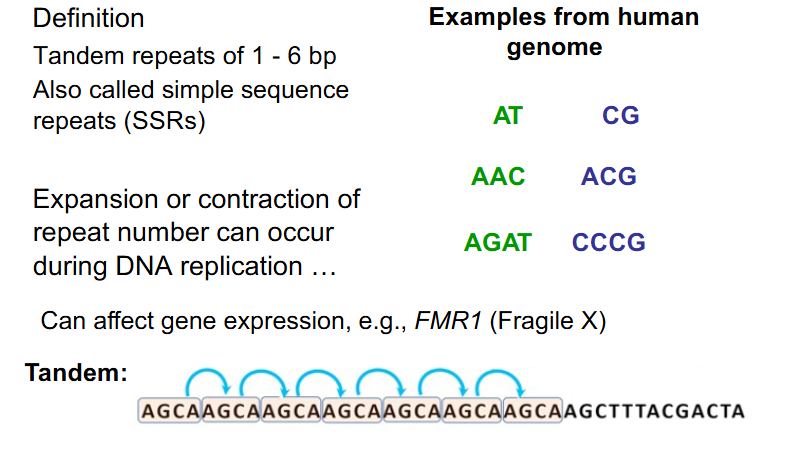
Repetitive Sequences - Microsatellites
- occur from DNA 1-100+ bp in length
- often occur directly adjacent to eachother (in tandem)
- also called simple sequence repeats (SSRs)
- Expansion or contraction of repeat number can occur during DNA replication
- can affect gene expression. e.g FMR1 (Fragile X)
- have to rmbr microsatellites as being tandem (adjacent to eachother) repeats of 1-6bp. microsatellites are also called SSRs
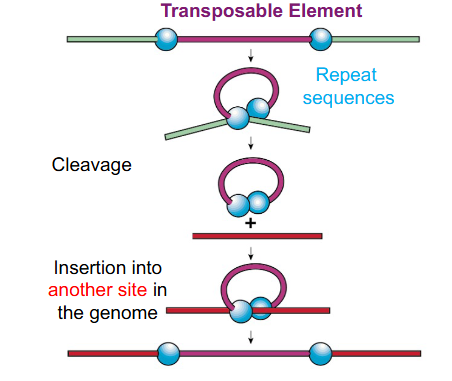
Transposable Elements
segments of DNA that can physically change their position within their genome
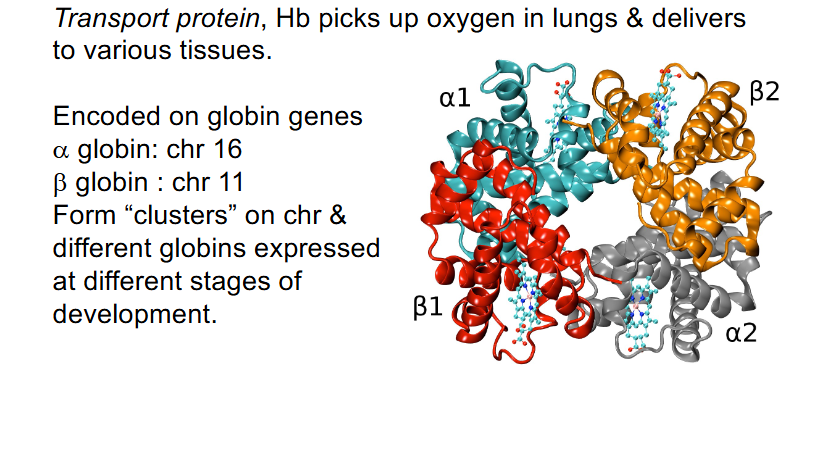
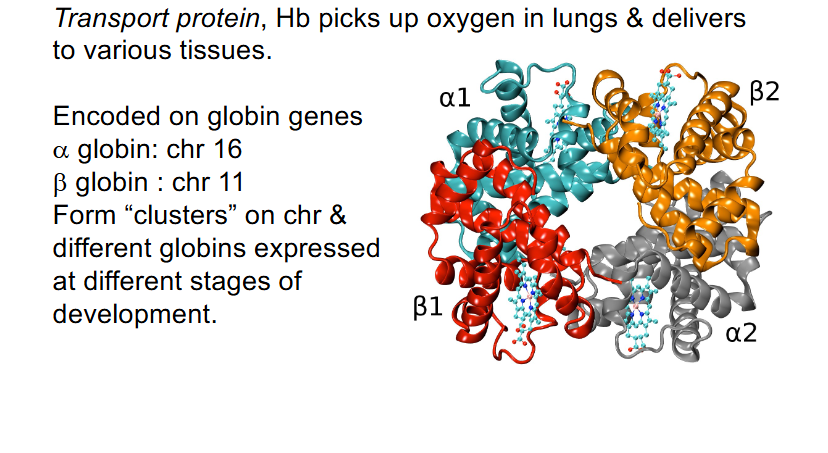
Haemoglobin (Hb): interesting genome structure
- Tetramer: 2 alpha and 2 beta polypeptide chains
- Transport protein: Hb picks up oxygen in lungs and delivers to various tissues
- encoded on globin genes: alpha globin: chr 16, beta globin: chr 11
- Form "clusters" on chr and different globins expressed at different stages of development
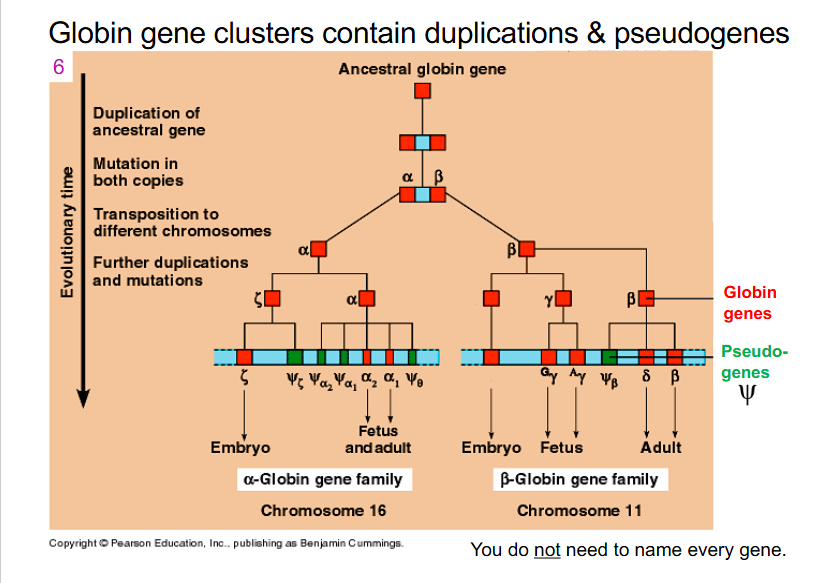
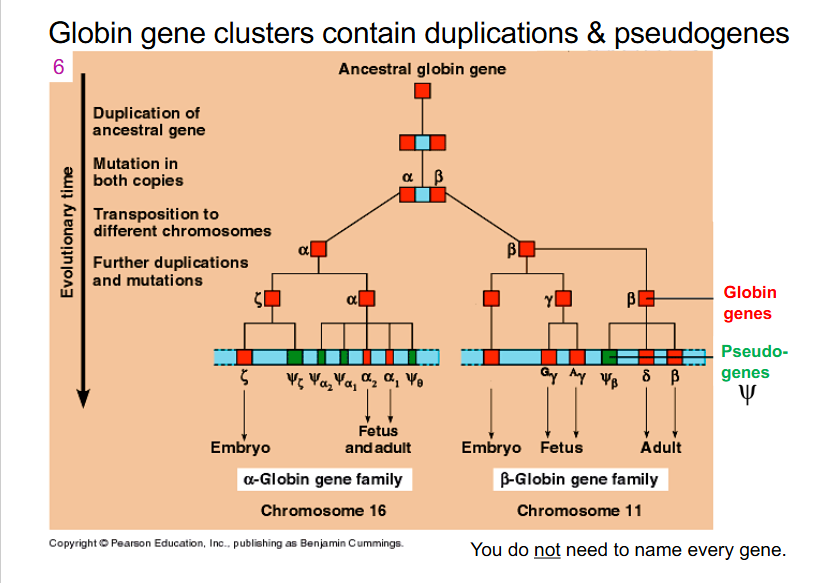
Globin gene clusters contain duplications and pseudogenes
- all the pseudogenes in slide do not maintain globin function, but they are found within our genome. can be retained and have evolved because we have copies of the functional alpha globin and beta globin genes as well
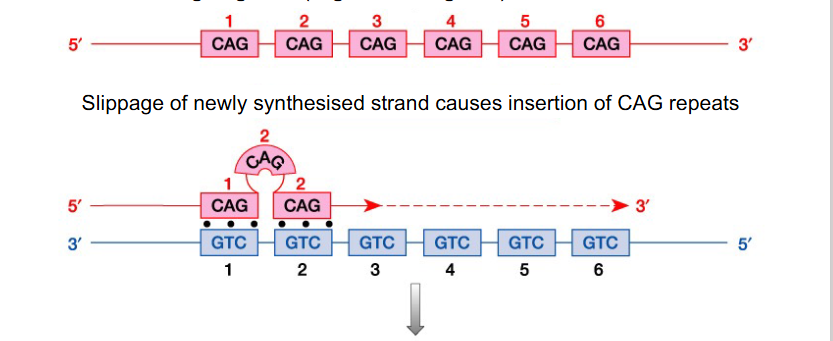
Replication Slippage
- repetitive sequences (i.e mini/micro satellites) can be anywhere in the genome and result from replication slippage
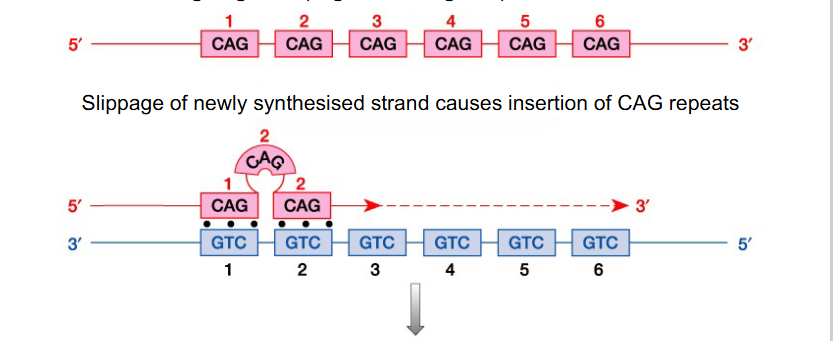
Repetitive Sequences - Minisatellites
- longer then microsatellites
- tandem repeats around 7-100bp long
- Also called VNTRs (variable number of tandem repeats)
- Highly variable in sequence length
- Can be in codinfg or non-coding regions
- Can affect gene expression
>1000 in human genome
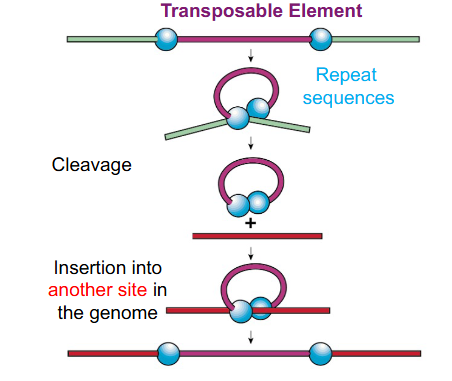
Transposable Elements Example
DIAGRAM ON SLIDE 24