Tertiary Structure Prediction
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
17 Terms
Nobel prize in physics
Fundamental algorithmic advances enabling machine learning with neural network
An example is backpropagation (popularised in part by Hinton), the algorithm trains neural networks by showing examples
Noble prize in Chemistry
Application of very advanced neural networks to predicting tertiary structure, e.g., AlphaFold
How to think about algorithms
Input→ Process → Output
Conceptually, can be multiple inputs and outputs
E.g., an algorithm to show the larger of two numbers
PSIPRED
A neural network
Position-specific scoring matrix
Trained through annotated examples via the PDB in 1999, probably many thousands
Output of DISOPRED
For each amino acid (x), a probability that it belongs to a disordered region is given
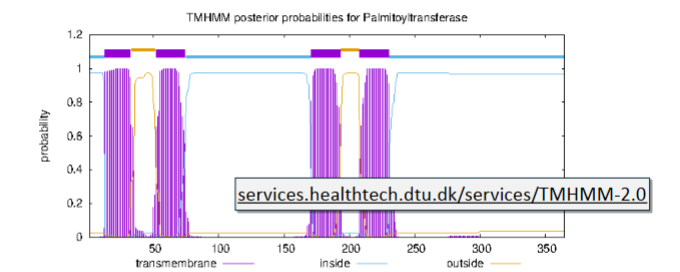
TMHMM: A hidden Markov model
Differs from neural networks: remembers context, called a state, as it processes each part of the sequence
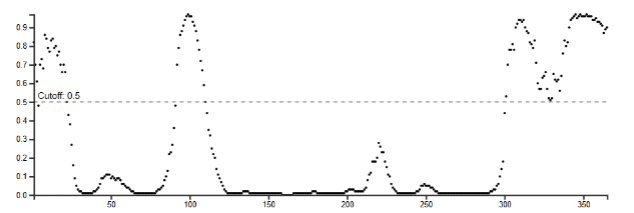
Output of TMHMM
For each amino acid (x), a transmembrane classification is given
Homology modelling
Search the PDB for the most statistically significant homologous sequence to the query, called the template
Bend the amino acid chain for the query such that each amino acid is the same position as its aligned counterpart in the template chain
Alignment of query and template
Every aligned residue used to match structures
Unaligned residues will be less accurate
Bending the chain with MODELLER
Long gaps, like this signal peptide, are not accurately modelled
Short gaps can be filled in accurately
Threading
As a rule of thumb, homology modelling works if there is a template with over 30% sequence identity
Threading, or fold recognition, is a method to model sequences in the twilight zone of low sequence identity
Generate models using a pool of possible templates, assess the plausibility of each attempt, and keep the best
Scoring functions for threading
PSSM similarity between query and template at each residue
Agreement of secondary structure prediction with template
Propensity of favourable side-chain interactions, e.g., hydrophobic-hydrophobic and polar-polar
Depth dependent structural alignment: looking across all the candidate templates, which of them share key structural features (like alpha helix in core or beta sheet at surface)
Sidechain packing
Possible sidechain orientations are given by a rotamer library, a statistical analysis of sidechains across the PDB
A good rotamer is common, doesn’t clash, and may make favourable interactions such as H-bonds
SCWRL4 is an automated method to choose good rotamers
AlphaFold
Produces highly accurate structures
AF3 accurately predicts structures across biomolecular complexes
Deep learning and attention
Means many hidden layers: these tend to each learn a different kind of information
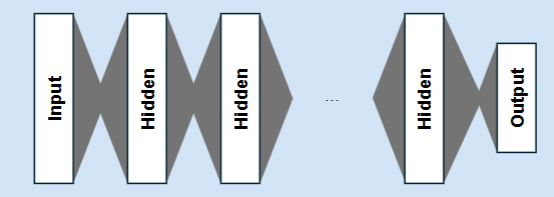
Attention
Allows the neural network to prioritise the most important information before each hidden layer
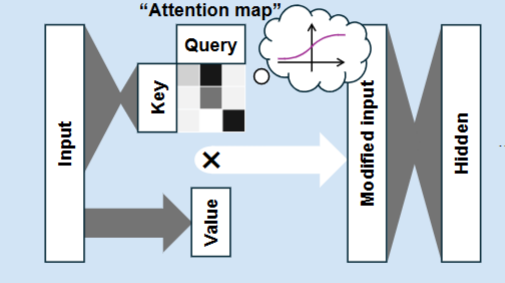
AlphaFold 3
A deep learning neural network with attention, which generates 3D models via diffusion