Chapters 21,22 - Translation and Mutations
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
What are special codons in translation?
During translation, there are start and stop codons, which are in charge of initiating or terminating/stopping this translation process.
Start codon: AUG, codes for methionine, which becomes the first amino acid in a protein’s chain.
Stop codons: UAA, UAG, UGA, do not code for any amino acids. When a ribosome encounters a stop codon, no tRNA (transfer RNA) with an anticodon can pair with it. Because of this, a release factor binds to the ribosome and causes it to release the polypeptide chain that was being made.
*Need to be able to read a codon chart
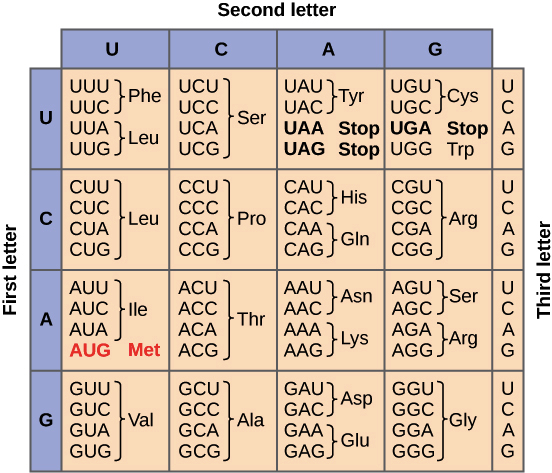
How does the linear sequence of bases in a messenger RNA corresponds to the primary structure of a polypeptide?
The linear sequence of bases in mRNA dictates the primary structure of a polypeptide in groups of three bases. These groups of three mRNA bases form a codon. During translation, each mRNA codon is read by the ribosome and each codon codes for a specific amino acid in the polypeptide chain. This is the polypeptide’s primary structure.
What is the relationship between the codons in a table and the DNA in the gene that corresponds to the codon? and what is the relationship between a codon and the anticodon loop in the tRNA?
The codons in a table are transcribed complementary to the DNA in a gene with the appropriate RNA bases (A,U,C,G, not T in RNA)
Ex: if our DNA that corresponds to a codon is TAC, it's transcribed mRNA codon is AUG.
The tRNA is in charge of recognizing the codon it has its anticodon for and translating to the corresponding amino acid. The tRNA’s anticodon is complementary to the mRNA codon.
Ex: if our mRNA codon is AUG, it’s corresponding tRNA has the anticodon UAC. This anticodon will code for methionine, which will be added to our polypeptide chain, starting the translation process. Overall, the mRNA codon AUG was translated to methionine.
Overall, the sequence of DNA bases in a gene determines the order of codons in mRNA (transcription). This mRNA sequence then determines the sequence of amino acids in a polypeptide chain (translation) leading to the creation of a unique protein.
Now, more in depth, what exactly is a tRNA?
In translation, tRNAs are transfer RNAs. Inside of a ribosome, they are in charge of recognizing the codons in an mRNA sequence and matching them to their corresponding amino acids; this is done through anticodons. Each tRNA carries an amino acid (if they don’t have an amino acid then they’re uncharged and won’t work) and has an anticodon sequence which binds to the mRNA codon it is complementary to. As a ribosome moves along an mRNA sequence, the amino acids that are obtained from each tRNA binding to mRNA codons are linked together forming a polypeptide chain.
Ex: the mRNA codon AAG will be recognized by the tRNA with the anticodon UUC (because it is complementary). The tRNA will then bind to the mRNA codon and code for the amino acid corresponding to AAG, lysine. This amino acid was carried by the tRNA.
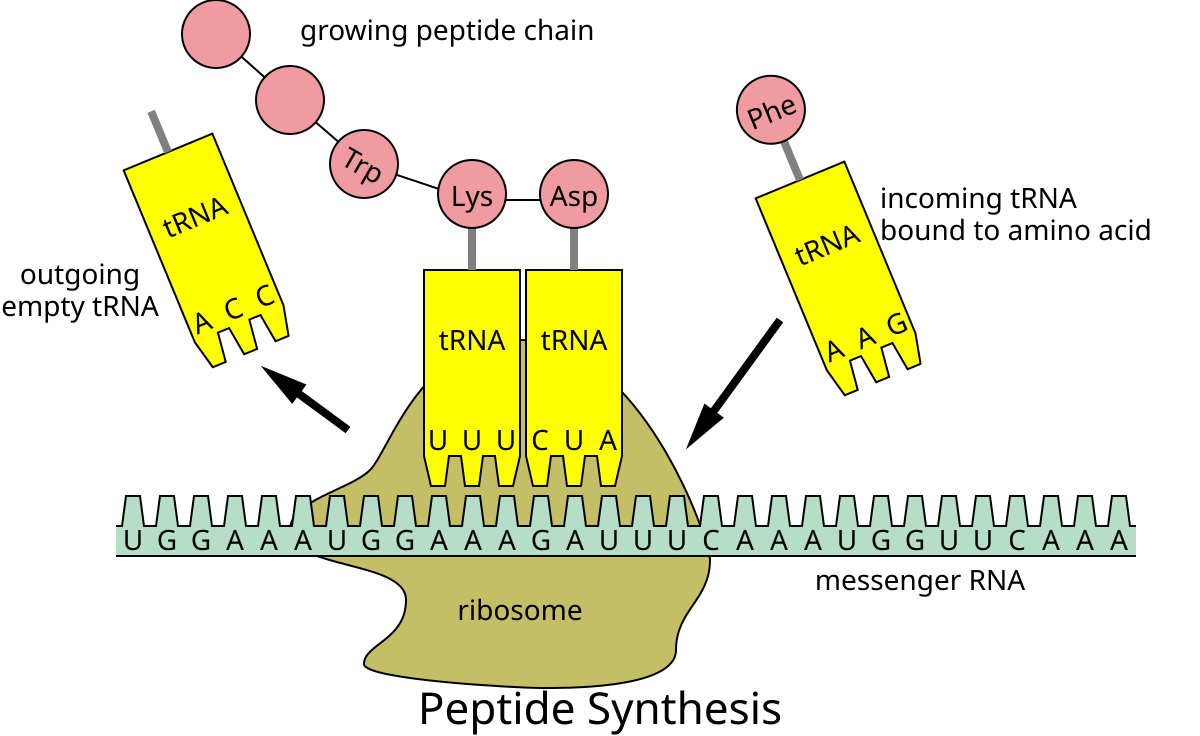
Describe the properties of ribosomes and how each are used during translation
Ribosomes are made up of two basic pieces: a large and a small subunit. These two subunits are composed of protein and rRNA (ribosomal RNA). During translation, these subunits come around/enclose an mRNA molecule (first the small subunit, then the large one) and move forward on the mRNA (5’ to 3’) reading codon by codon (groups of 3 bases); this is known as translocation. After translation is finished (a protein is built) these two subunits separate allowing for a ribosome to be reused.
Ribosomes also have three different slots/sites: the A site, the P site, and the E site (from right to left). These are used by the tRNAs to move along during translation.
A site: the first site a tRNA moves through. In here, the tRNA, charged with its corresponding amino acid, enters the ribosome. The tRNA’s anticodon is matched to the proper mRNA codon sequence.
P site: In here, the tRNA in the P site holds the growing polypeptide chain. When a new charged tRNA enters the A site, a peptide bond forms between the amino acid on the P-site tRNA and the one on the A-site tRNA. After the bond forms, the P-site tRNA becomes uncharged and moves to the E site (its corresponding mRNA codon sequence also moves), while the tRNA that was in the A site now carries the growing chain and shifts into the P site as the mRNA moves forward. This process keeps on going as the mRNA sequence continue to be read.
E site: Also known as the exit site. The uncharged tRNA that used to be in the P site is now in the E site. In here, such tRNA detaches from the mRNA codon sequence and leaves the ribosome through this E site. The mRNA sequence moves along (5’ to 3’) and the ribosome reads the next mRNA codon.
Important to remember: in eukaryotes, some ribosomes are wandering free in the cytosol and other are bound to the rough ER, it depends on whether the protein is meant to be secreted or for membranes.
In prokaryotes, the ribosomes are in the cytosol.
Another note: several ribosomes can translate the same mRNA forming a polyribosome (polysome).
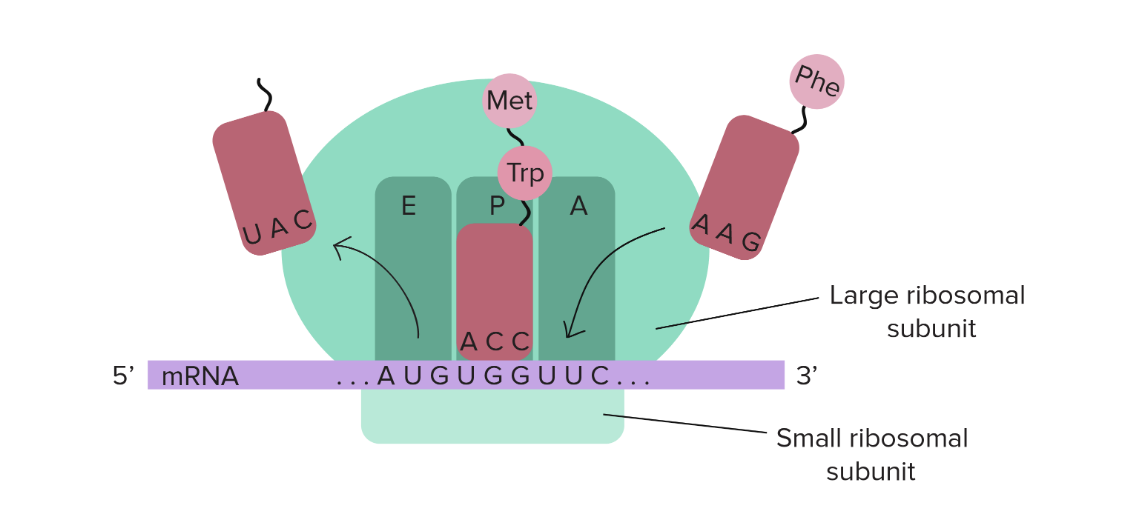
Explain the steps of translation during the ribosome cycle, including the three steps of the elongation cycle and the source of energy.
During translation, three steps take place inside the ribosome: initiation, elongation and termination.
Initiation: the small subunit of the ribosome is built around the mRNA sequence we want to read. As the mRNA sequence is read, the starting codon AUG (which codes for methionine) will eventually be reached. Once the tRNA with the proper anticodon and the AUG codon are attached, translation begins and the larger subunit of the ribosome joins; the mRNA is now fully enclosed by the ribosome.
Elongation: the tRNA that is attached to an mRNA codon moves along the ribosome and the mRNA codon also moves but because it is read by the ribosome. As the tRNA enters the A site, its amino acid becomes the first in the polypeptide chain through a polypeptide bond. The amino acid is then released in the P site and the tRNA moves along to the E site (because without an amino acids is now uncharged) where it is released.
The next tRNA enters the A site and as its amino acid is added to the polypeptide chain through a peptide bond, the growing polypeptide chain is now on this tRNA. This tRNA lets go of these in the P site, moves to the E site to be released and this cycle continues until a stop codon is read.
Termination: once a stop codon is read, a release factor enters the ribosome’s A site. Through hydrolysis (the addition of a water molecule to break a bond), these release factors cause the last tRNA (with the anticodon corresponding to the mRNA codon that is not the stop one) to release the polypeptide chain created. A protein is released from the ribosome and the ribosome subunits separate.
Overall: energy comes from GTP (all ribosome movement) and ATP hydrolysis (for charging the tRNAs with their corresponding amino acid before they enter the A ribosomal site)
Major differences between translation in eukaryotes and prokaryotes.
Prokaryotes have no nucleus, therefore: transcription and translation (which happens inside a ribosome) occur in the cytosol (liquid of the cytoplasm). These processes can occur simultaneously.
Eukaryotes do have a nucleus and membrane-bound organelles, therefore: transcription occurs in the nucleus and translation in the cytoplasm (the ribosomes are either wandering around in the cytosol or attached to the rough ER). Also remember that transcription in eukaryotes has the three extra editing steps. Because of this, transcription and translation may not occur simultaneously.
What is mutation?
A heritable change in the genetic material. Mutations result from changes in the DNA sequence and usually affect the tertiary structure of a protein and can therefore sometimes affect its overall function.
What are some common causes of mutation?
Mutations occur as the result of changes in the DNA sequence. These changes are often caused by spontaneous or induced reasons:
Spontaneous mutation: mutation results from natural errors in DNA replication or repair, or from errors in DNA due to chemical changes (very rare).
Induced mutation: mutation results from exposure to external agents, known as mutagens, that damage to DNA and fail to repair such. This is more frequent than spontaneous mutation and some examples are x-rays, uv-lights, chemical mutagens, etc.
How are mutations passed from generation to generation?
In single-celled organisms: mutations are passed to the next generation
In multi-cellular organisms: passed through one of two ways:
Somatic mutation: mutations in somatic cells (all body cells besides sex cells) are not passed on and only affect the current carrier individual.
Germline mutation: mutations on sperm or eggs (in gametes, sex cells). These are passed on to the next generation.
Why doesn’t a translation machinery detect mutations
Ribosome reads codons blindly; it cannot “know” whether protein sequence is normal.
RNA polymerase and ribosomes simply follow base-pairing and codon rules, they copy and read whatever nucleotide sequence is present. Because of this, if the DNA or mRNA carries a mutation, the machinery (the RNA polymerase and ribosomes) will still transcribe and translate it as usual, producing a mutated mRNA and possibly an altered protein. The cell does not “know” that the sequence is incorrect.
What are the types of mutation?
Aneuploidy: changes in chromosomal number. Ex: down syndrome (trisomy 21, three chromosomes on pair 21).
Large-scale mutation: large-scale changes in chromosomes such as insertions, translocations, or large deletions of chromosome segments. These can affect several genes at once.
Point mutations: small DNA changes within a gene. These can be silent, missense, nonsense, or frameshift and can result from substitution of a single base (the overall number of bases stays the same), or from an insertion or deletion (the overall number of bases changes); these mutations may or may not lead to changes in a protein’s amino acid sequence (specifics will be discussed further).
What are the different types of base substitutions in a coding region? and what are their effects on the resulting protein?
Base substitutions involve the substitution of a single nucleotide (in DNA) without changing the overall number of bases in the DNA or mRNA sequence. There are 3 main types:
silent mutation: a base (a letter) is changed in the DNA sequence leading to a base change in the mRNA sequence and therefore a change on a codon (a sequence of 3 bases). However, this codon still codes for the same amino acid it used to code before the mutation. Because of this, no changes occur to the final protein.
Ex: CAA (glutamine) is altered to CAG (also glutamine); therefore, no change!
missense mutation: a base (a letter) is changed in the DNA sequence leading to a base change in the mRNA sequence and therefore a change on a codon (a sequence of 3 bases). This codon now codes for a different amino acid that the one it used to code for before the mutation. Because of this, the amino acid sequence changes and therefore the protein’s structure and function (we obtain a different protein).
Ex: CAA (glutamine) is altered to CCA (proline); therefore, there is a change!
nonsense mutation: a base (a letter) is changed in the DNA sequence leading to a change in the mRNA sequence and a STOP codon. This leads to a premature stop codon because it is placed before the original stop codon. Translation of the mRNA sequence is terminated early and therefore, the protein obtained is now shorter than what it was supposed to.
Ex: CAA (glutamine) is altered to UAA (stop codon);
What are insertion or deletion mutation?
The mutations where there are DNA changes (just like in all mutations) but nucleotides (contains a base) are added or removed.
This alters the overall number of bases in the DNA and mRNA sequences. By inserting or deleting a base (a letter) in DNA, the mRNA sequence changes and so does the way the codons are read (a codon consists åof 3 bases). This is known as a frameshift!
This frameshift leads to different amino acids being coded for (because we’re now reading different codons) and therefore a different protein structure.

Lastly, how can mutations change protein function?
Mutations lead to changes in the DNA sequence of a gene, which can alter the mRNA transcribed from it. This may lead to the bases of an mRNA sequence to be changed and therefore lead to changes in the amino acids. This then affects protein structure (folding, and then 3D shape) and therefore function.
Overall: DNA - mRNA - amino acid sequence - protein folding (structure) - function.