Biochemistry I Lesson 3: Enzyme Kinetics
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
Write the rate law for the following reaction: A+B -> AB
Rate= k [A][B]
Rate is the change in concentration per unit time.
k is the rate constant (depends on the reaction).
[A] and [B] are the concentrations of reactants A and B.
*Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/
In Michaelis-Menten Kinetics experiments, which of the following is assumed to be true?
I. Substrate concentration is constant.
II. Increasing the Enzyme concentration will not change Vmax.
III. The enzymes are saturated at Vmax.
(A) I Only
(B) III Only
(C) I and II Only
(D) I, II, and III
(B) III Only
In Michaelis-Menten Kinetics experiments:
- ENZYME concentration is held constant as we increase the substrate concentration.
- If you were to increase the enzyme concentration, the Vmax would increase.
- The enzymes are saturated at Vmax.
In Michaelis-Menten Kinetics experiments, which of the following is assumed to be true?
I. Our Solutions are behaving ideally.
II. Our constants ([E] and k) are not changing during the experiment.
III. Substrate can be converted into product with or without the enzyme.
(A) I Only
(B) III Only
(C) I and II Only
(D) I, II, and III
(C) I and II Only
In Michaelis-Menten Kinetics experiments, we assume that:
- Our Solutions are behaving ideally.
- Our constants ([E] and k) are not changing during the experiment.
- Substrate CANNOT be converted into product without the enzyme.
How can we increase the rate of a reaction assuming the rate constant (k) is constant?
I. Increase Substrate Concentration
II. Increase Enzyme Concentration
III. Increase Mixed Inhibitor Concentration
(A) I Only
(B) I and II Only
(C) II and III Only
(D) I, II, and III
(B) I and II Only
We can increase the rate of reaction by increasing the substrate or enzyme concentration.
Adding any type of inhibitor will not increase the rate of a reaction.
True or false? Adding more catalyst after a catalytic amount has already been added will increase the rate of reaction.
False. Adding more catalyst after a catalytic amount has already been added will NOT AFFECT the rate of reaction.
You only need a small amount ("a catalytic amount") of catalyst for it to do its job. Adding more will not do anything.
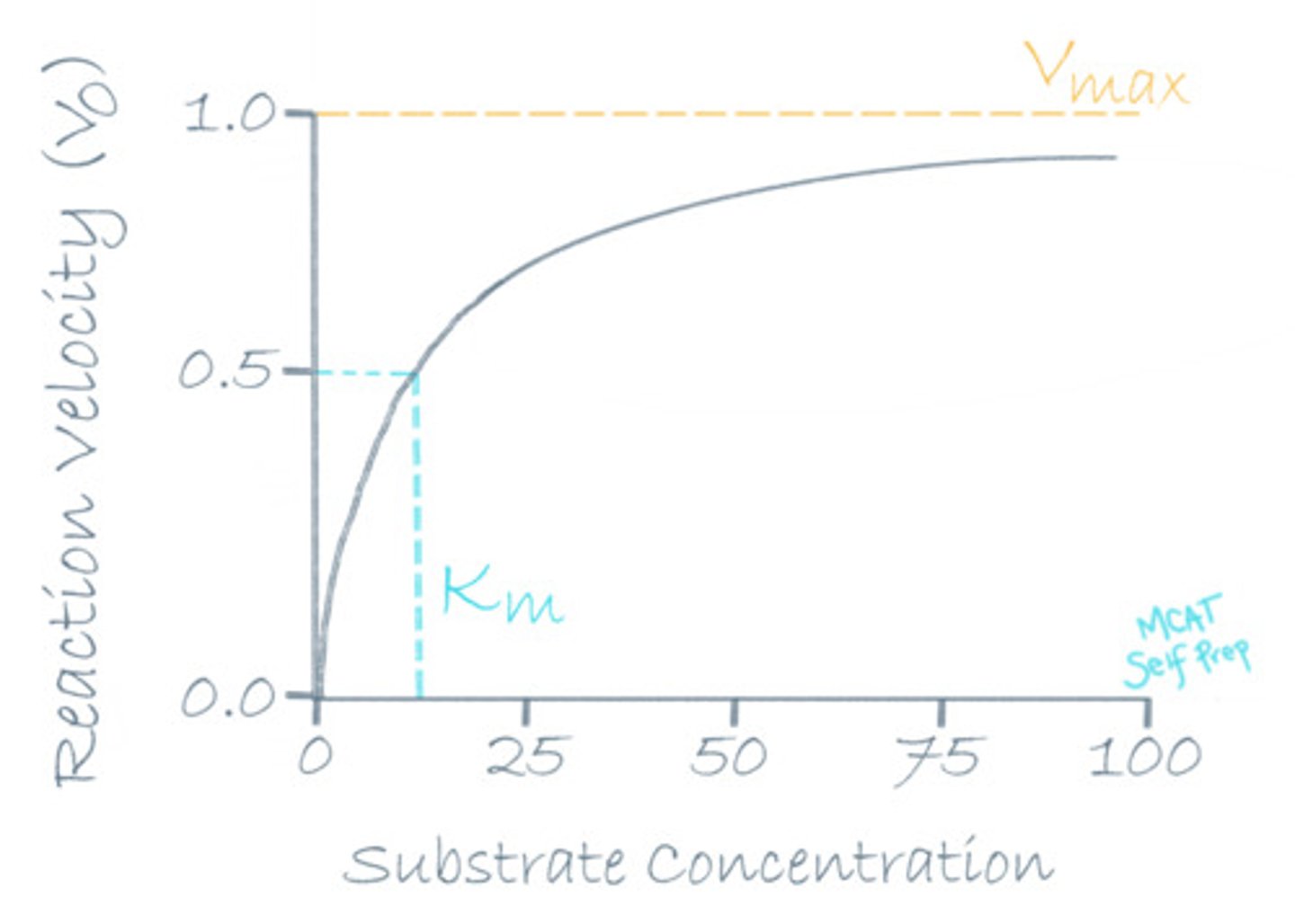
What does it mean if the rate of reaction has reached maximum velocity (Vmax)?
(A) There are more products than reactants.
(B) The substrates are no longer able to get any closer to the enzymes.
(C) The substrates are no longer able to get in any better of an orientation.
(D) The enzymes no longer have available active sites.
(D) The enzymes no longer have available active sites.
When the rate of reaction reaches Vmax, it means that the enzymes are saturated and will not be able to react any more quickly since all active sites are filled up with substrates.
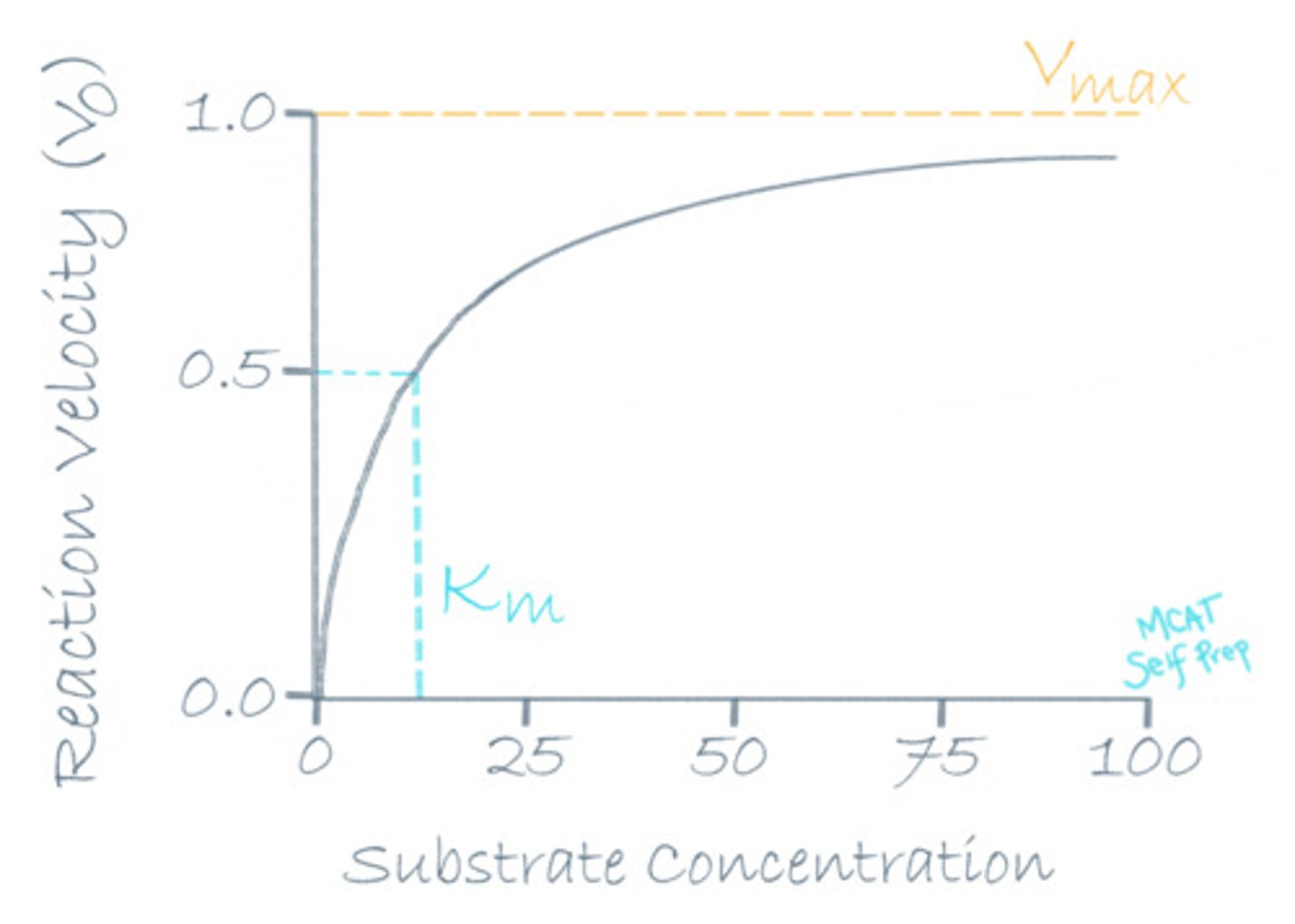
If a reaction has reached Vmax, what would happen to the rate of reaction if we increase the substrate concentration?
(A) It would increase
(B) It would remain the same
(C) It would decrease
(D) It would stop
(B) It would remain the same
Increasing the substrate concentration would have no effect on the rate of the reaction because all active sites are already occupied at Vmax.
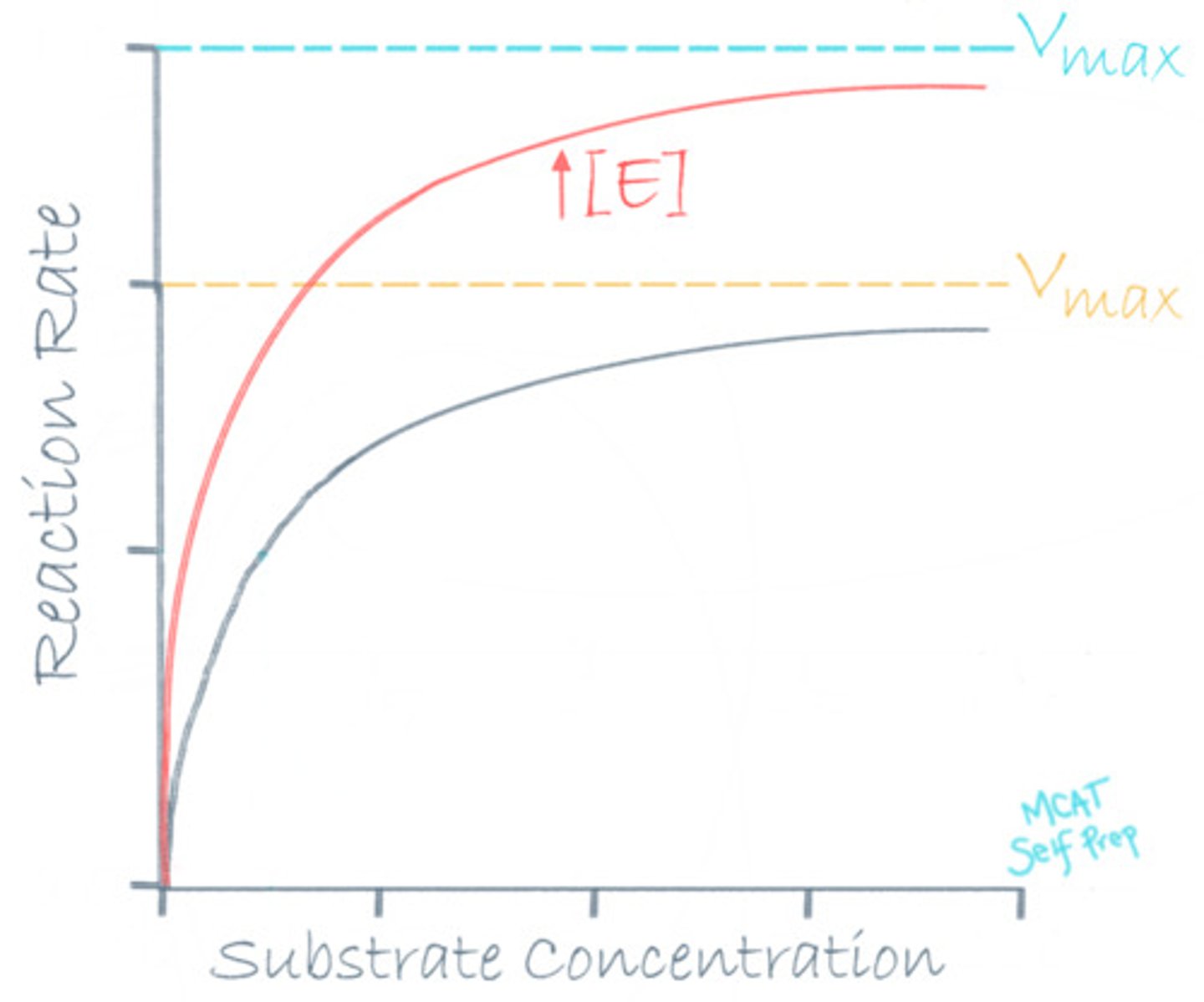
What is the one way to increase the Vmax of a reaction?
(A) Increase Substrate Concentration
(B) Increase Enzyme Concentration
(C) Increase Inhibitor Concentration
(D) Increase Product Concentration
(B) Increase Enzyme Concentration
The only way to increase Vmax is by increasing the enzyme concentration.
These are the two steps of enzyme catalysis:
E + S => ES => E + P
What are the respective reaction rate formulas?
E + S => ES => E + P
Rate1 = k1 [E][S]
Rate2 = k2 [ES]
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/
![<p>E + S => ES => E + P</p><p>Rate1 = k1 [E][S]</p><p>Rate2 = k2 [ES]</p><p>Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/</p>](https://knowt-user-attachments.s3.amazonaws.com/2d1bef02-9f11-4b44-b8e3-c9839a799783.jpg)
What is the steady-state assumption when talking about enzyme kinetics?
(A) [ES] is constant
(B) [S] is constant
(C) [P] is constant
(D) [I] is constant
(A) [ES] is constant
The steady state assumption means that the concentration of the enzyme-substrate complex (ES) is constant, which means that the formation of ES is equal to the dissociation of ES.
Write out the Michaelis-Menten equation. What do each of the variables mean?
The Michaelis-Menten equation is a very important equation when talking about enzyme kinematics and is shown in the picture.
Don't worry about knowing how to derive this equation. Just memorize it and understand each variable.
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/

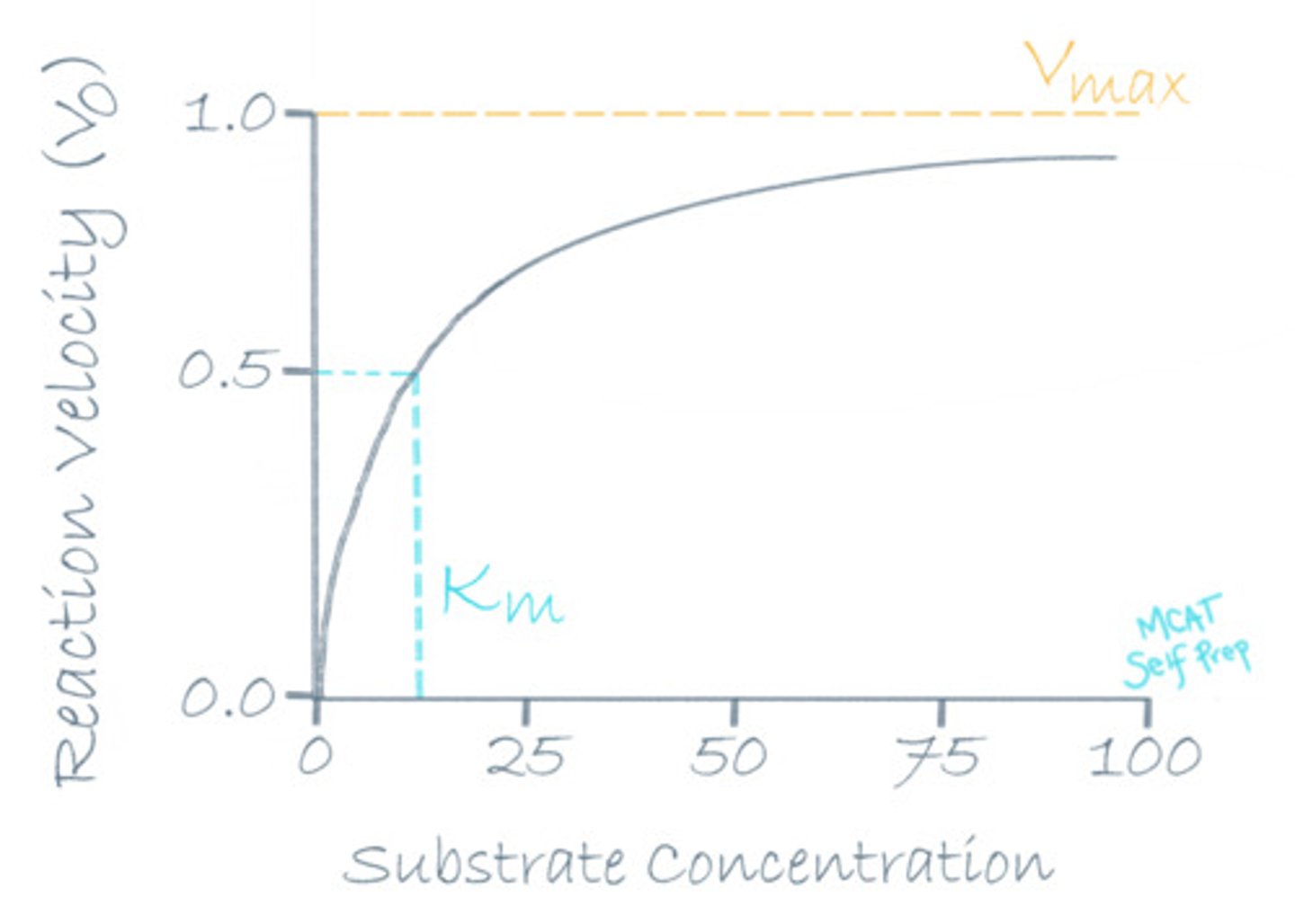
What is the Michaelis constant (Km)?
(A) The Vo at half of Vmax
(B) The Vmax at half of Vo
(C) The [S] at half of Vmax
(D) The Vmax at half of [S]
(C) The [S] at half of Vmax
The Michaelis constant (Km) is the substrate concentration where the initial velocity (Vo) is equal to 1/2 of Vmax.
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/
![<p>(C) The [S] at half of Vmax</p><p>The Michaelis constant (Km) is the substrate concentration where the initial velocity (Vo) is equal to 1/2 of Vmax.</p><p>Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/</p>](https://knowt-user-attachments.s3.amazonaws.com/71059388-c42e-4500-9e22-420dc528a4a6.png)
The catalytic efficiency for a certain reaction increases. What happens to the reaction rate?
(A) It would increase
(B) It would remain the same
(C) It would decrease
(D) It would stop
(A) It would increase
Catalytic efficiency is basically an enzyme's ability to catalyze reactions. If it increases so too will the rate of the reaction.
What is the equation for the enzyme turnover number (Kcat)?
Kcat = Vmax/ [E]T
Vmax is the maximum velocity of the enzyme
[E]T is the concentration of the enzyme.
unit: 1/sec
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/
![<p>Kcat = Vmax/ [E]T</p><p>Vmax is the maximum velocity of the enzyme</p><p>[E]T is the concentration of the enzyme.</p><p>unit: 1/sec</p><p>Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/</p>](https://knowt-user-attachments.s3.amazonaws.com/3ebf6d4c-3e9c-43ac-92a6-d1db03ec12bc.jpg)
What does the enzyme turnover number (Kcat) really mean?
The enzyme turnover number (Kcat) basically tells us how many substrates a single enzyme can turn into product in one second at its maximum speed.
CRB True or false? Kcat values between enzymes are somewhat constant, with many having between 101 and 103 products per enzyme per second as their Kcat.
True. Kcat values between enzymes are somewhat constant, with many having between 101 and 103 products per enzyme per second as their Kcat.
What is the formula for catalytic efficiency?
Catalytic Efficiency = Kcat/Km.
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/

The enzyme pepsin is unable to break down proteins into smaller peptides thus causing a digestion problem in the stomach. What can we do to increase pepsin's catalytic efficiency?
I. Increase Km
II. Increase Kcat
III. Increase Vo
(A) I Only
(B) II Only
(C) I and II Only
(D) I, II, and III
(B) II Only
To increase catalytic efficiency, we need to increase the enzyme turnover number (Kcat) or DECREASE the Michaelis constant (Km).
True or False? If you wanted to compare which enzymes are better at speeding up reactions, you could compare the catalytic efficiency of the enzymes.
True. You can rate enzymes by calculating their catalytic efficiency, which is (Kcat/Km); the higher the catalytic efficiency, the better the enzyme is at speeding up reactions.
Draw a Lineweaver-Burke plot. What is the equation for the x- and y- intercept? What about the slope?
Struggling to keep your MCAT equations straight? Simply conquer the 100 most important equations using Andrew's 100 Most Essential Equations Mastery Course @ https://mcatselfprep.com/course/andrews-equation-mastery-course/
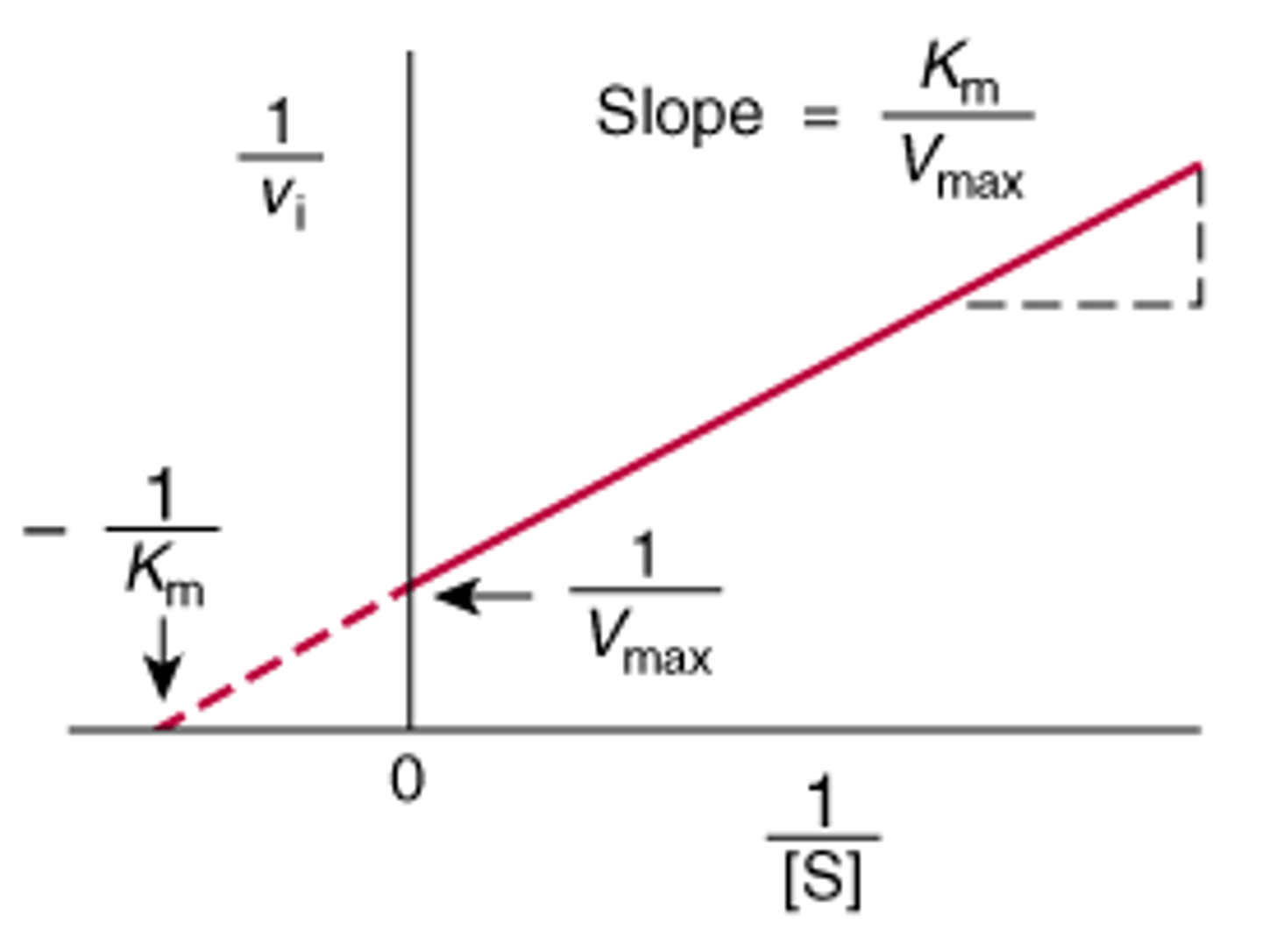
CRB Which of the following is NOT typically and easily identified using Lineweaver-Burke Plots?
(A) Km
(B) Kcat
(C) Vmax
(D) The type of inhibition an enzyme faces.
(B) Kcat
Lineweaver-Burke Plots are often used to identify Km, Vmax and the type of inhibition an enzyme is facing.
What are the similarity(s) and difference(s) between the following:
(1) Competitive inhibitors
(2) Uncompetitive inhibitors
(3) Non-competitive inhibitors
(4) Mixed inhibitors
Competitive inhibitors bind to directly to the enzyme at the active site (they compete for the active site!).
Uncompetitive inhibitors bind at an allosteric site of the enzyme while the substrate is already bound to the enzyme (UN = ES, 2 letters on each side).
Non-competitive inhibitors have equal affinity for the enzyme and the enzyme-substrate complex (NON = E or ES, 3 letters on each side).
Mixed inhibitors have different affinity for the enzyme and enzyme-substrate complex (just like NON, but it is uneven/mixed).
A patient is rushed into the ER due to methanol poisoning, in which methanol is enzymatically converted to toxic metabolites. The doctor treats the patient by administering intravenous ethanol to compete for the active sites of the enzymes involved to prevent the production of toxic metabolites. What type of inhibitor is this?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(A) Competitive
We should know this is a competitive inhibitor because the inhibitor works by binding to the active site of the enzyme.
An inhibitor binds to a free enzyme preventing it from being able to react with the substrate. That same inhibitor is also able to bind to the enzyme-substrate complex, preventing the enzyme from turning the substrate into product. What type of inhibitor could this be?
I. Uncompetitive
II. Noncompetitive
III. Mixed
(A) I Only
(B) II Only
(C) I and II Only
(D) II and III Only
(D) II and III Only
This can be either a non-competitive inhibitor or mixed inhibitor because it can bind to either the enzyme or the enzyme-substrate complex.
Uncompetitive inhibitors only bind to the ES complex.
Which type of inhibitor will decrease Vmax but does not apparently alter the value of Km?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(C) Noncompetitive
A noncompetitive Inhibitor will decrease Vmax and not alter the value Km.
A noncompetitive inhibitor decreases Vmax because it binds to enzyme and the enzyme-substrate complex, thus creating less available enzymes to react.
The inhibitor binds ES, shifting the reaction (E + S --> ES) to the right. The inhibitor also binds to E, shifting the reaction to the left. Furthermore, the inhibitor binds to ES and E with the same affinity; this means these shifts cancel each other out, resulting in no change to affinity (Km).
Note that this is NOT what Khan Academy says. Their description is incorrect!
Which type of inhibitor decreases Vmax and can either increase or decrease Km?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(D) Mixed
A mixed inhibitor always decreases Vmax; but can either apparently increase or decrease Km (decrease or increase affinity for substrate) since a mixed inhibitor has different affinities for the enzyme and enzyme-substrate complex.
If a mixed inhibitor prefers binding to the enzyme, the Km will increase.
If mixed inhibitor prefers binding to the enzyme-substrate complex, the Km will decrease.
Which type of inhibitor will increase Km without affecting the Vmax?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(A) Competitive
Competitive inhibitors will increase Km (decrease affinity for the substrate), but leave Vmax unchanged because if enough substrate is added, it will outcompete the inhibitor and the reaction will still run at maximum velocity.
Which type of inhibitor will always decrease both Km and Vmax?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(B) Uncompetitive
Uncompetitive inhibitors will apparently decrease Km (increase substrate affinity). This is because binding to ES will shift the reaction (E + S --> ES) to the right (the forward direction).
These inhibitors will also decrease Vmax because they bind to the enzyme-substrate complex, preventing these bound enzymes from completing any further reactions. This essentially decreases the concentration of E, decreasing Vmax.
Note that a noncompetitive mixed inhibitor can also decrease both Vmax and Km if it binds more to the ES complex than to E alone. However, not all mixed inhibitors are like this, so it is not the best answer.
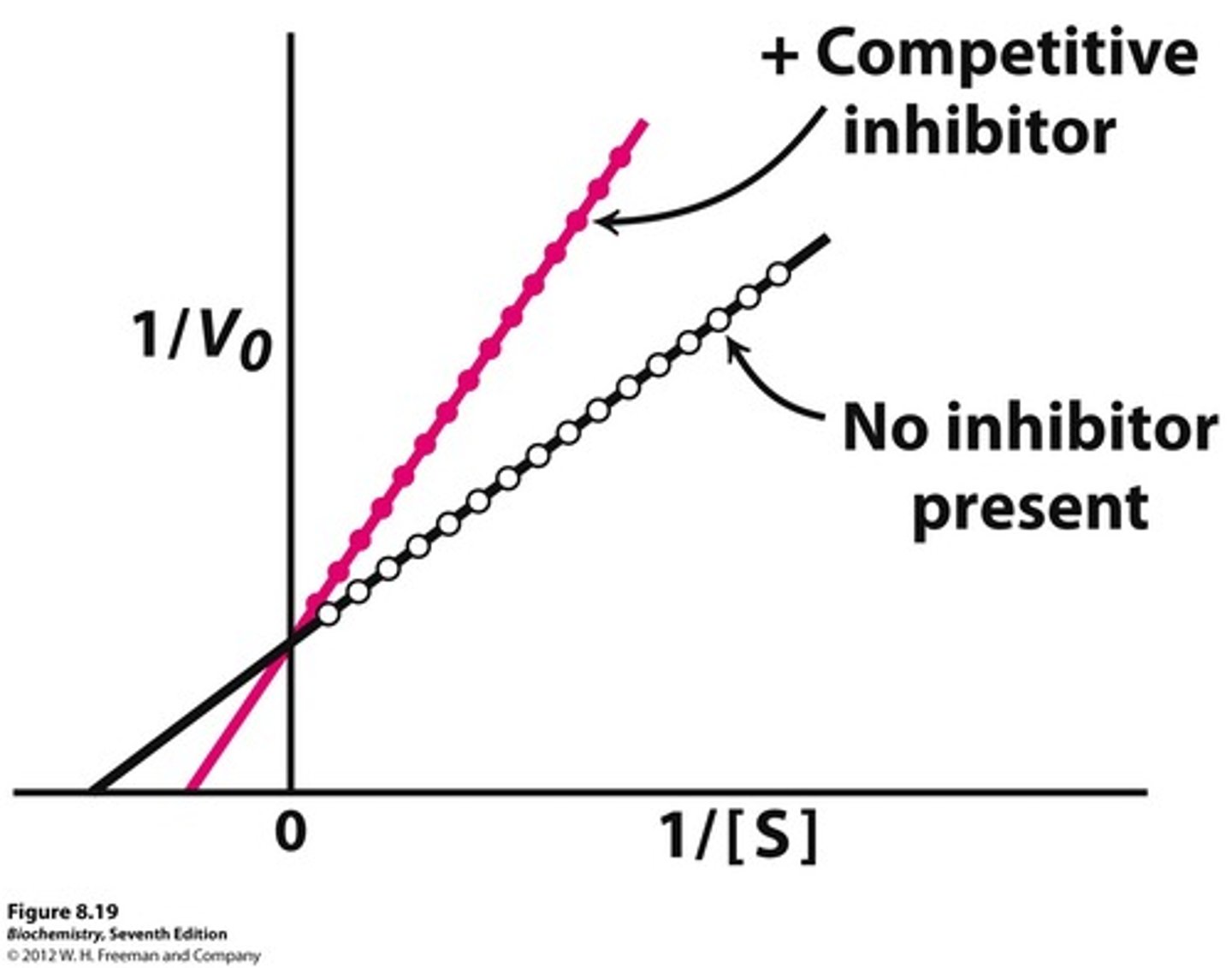
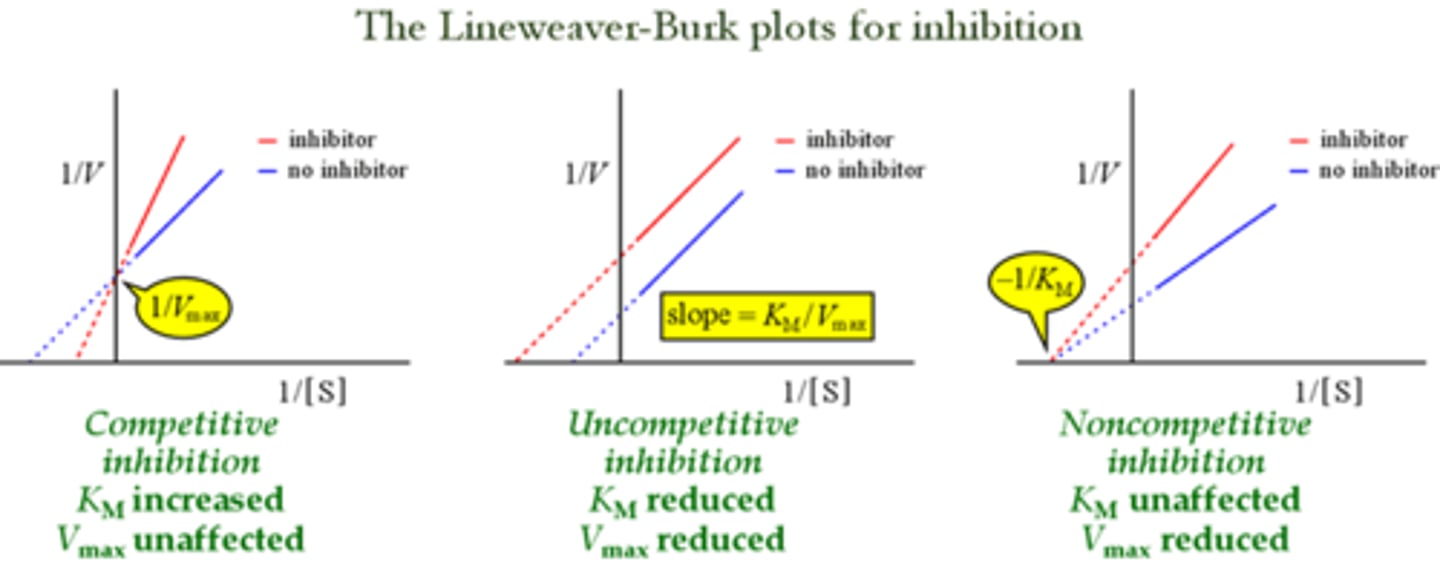
Describe, or draw, how the Lineweaver-Burke plot will look for a competitive inhibitor compared to the uninhibited enzyme.
The Lineweaver-Burke plot for a competitive inhibitor will have the same y-intercept as the uninhibited enzyme, but a less negative x-intercept.
This illustrates that the Km increased, but the Vmax did not change.
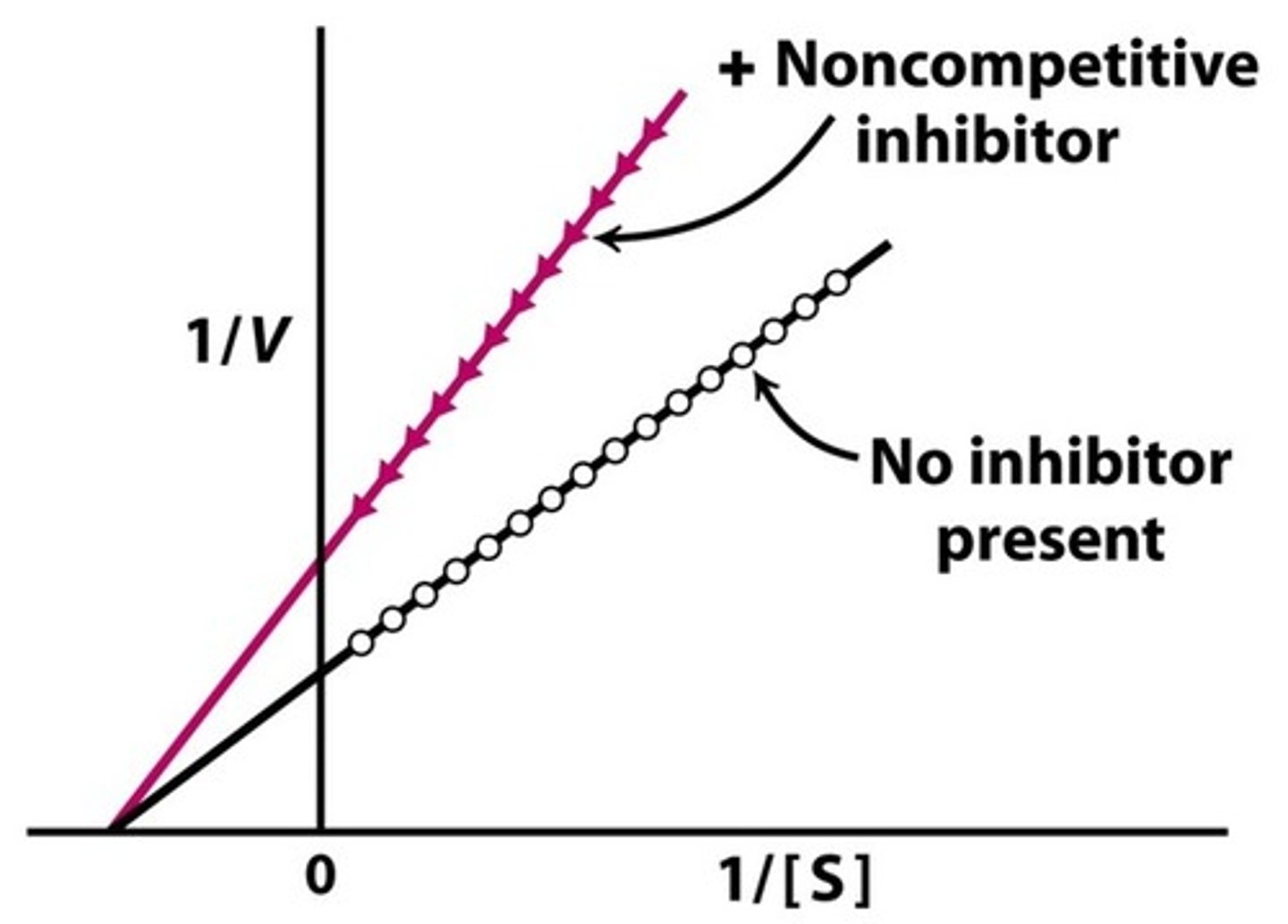
Describe, or draw, how the Lineweaver-Burke plot will look for a non-competitive inhibitor compared to the uninhibited enzyme.
The Lineweaver-Burke plot for a noncompetitive inhibitor will have the same x-intercept, but a higher y-intercept and slope. This indicates that the Km did not change, but the Vmax decreased.
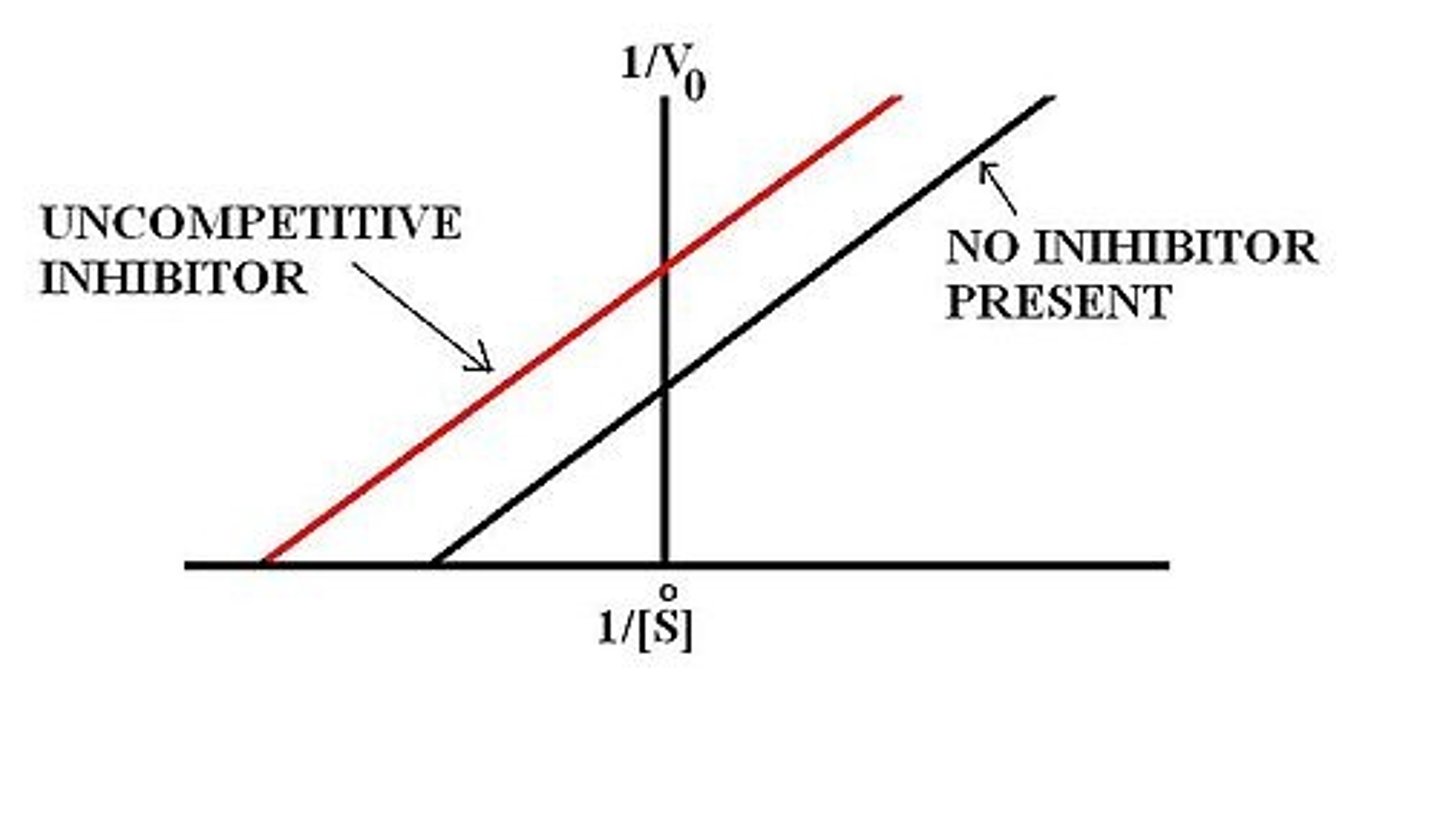
Describe, or draw, how the Lineweaver-Burke plot will look for an uncompetitive inhibitor compared to the uninhibited enzyme..
The Lineweaver-Burke plot for an uncompetitive inhibitor will be parallel to the uninhibited enzyme's plot with a higher y-intercept and a more negative x-intercept. This indicates a decreased Vmax and a decreased Km.
CRB To further highlight the differences between Competitive, Uncompetitive and Noncompetitive Inhibition, draw all three of their Lineweaver-Burke plots side-by-side.
Describe, or draw, how the Lineweaver-Burke plot will look for a mixed inhibitor with respect to no inhibitor.
The Lineweaver-Burke plot for a mixed inhibitor will have an increased y-intercept and an increased or decreased x-intercept.
Increasing the substrate concentration will only be able to overcome the inhibitory effects of what kind of inhibitor?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(A) Competitive
Increasing the substrate concentration will only be able to overcome the inhibitory effects of competitive inhibitors.
What type of inhibitor does not change the apparent Km value?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(C) Noncompetitive
A noncompetitive inhibitor does not change the apparent Km value because the inhibitor binds equally well to the enzyme and enzyme-substrate complex.
What type of inhibitor does not change the Vmax value?
(A) Competitive
(B) Uncompetitive
(C) Noncompetitive
(D) Mixed
(A) Competitive
Competitive inhibitor does not change the Vmax value because if enough substrates are added, it will outcompete the inhibitor and be able to run the reaction at maximum velocity.
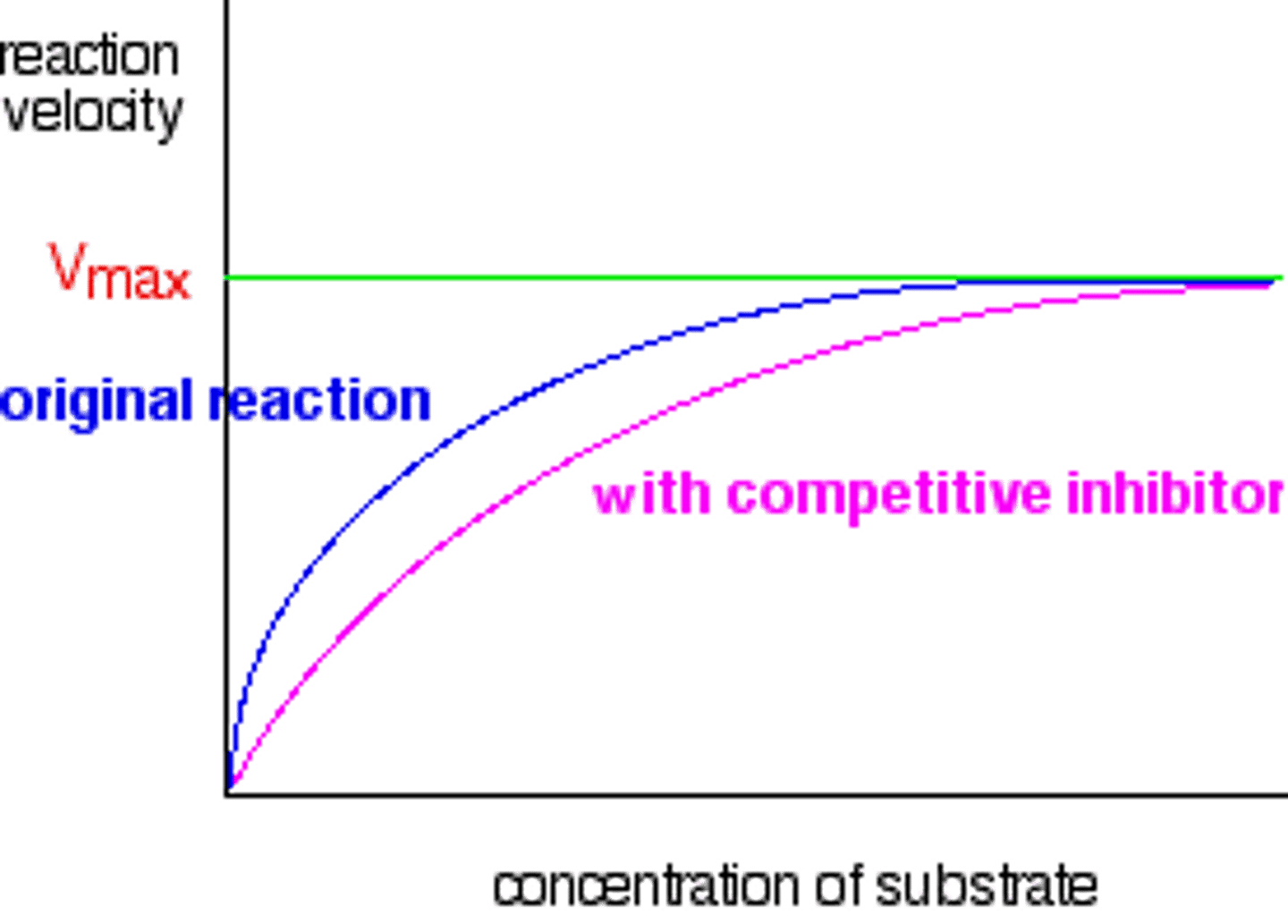
What is enzyme cooperativity?
(A) Substrate binding that changes substrate affinity
(B) Substrate affinity that changes substrate binding
(C) Enzyme binding that changes product affinity
(D) Product binding that changes Enzyme affinity
(A) Substrate binding that changes substrate affinity
Enzyme cooperativity is substrate binding that changes substrate affinity.
Substrate binding that does not affect the affinity for subsequent substrates is known as:
(A) positive-cooperative binding.
(B) negative-cooperative binding.
(C) neutral-cooperative binding.
(D) non-cooperative binding.
(D) non-cooperative binding.
It is called non-cooperative binding since the substrate binding does not affect the affinity for subsequent substrates.
What is the difference between positive-cooperative binding vs negative-cooperative binding?
Positive-cooperative binding is when substrate binding INCREASES the affinity for subsequent substrates.
Negative-cooperative binding is when substrate binding DECREASES the affinity for subsequent substrates.
On a Michaelis-Menten plot please draw what the curves would look like for enzymes with non-cooperative binding, positive-cooperative binding, and negative-cooperative binding.
An enzyme with non-cooperative binding will have a hyperbolic shaped curve.
An enzyme with positive-cooperative binding will have a sigmoidal "S" shaped curve.
An enzyme with negative-cooperative binding will have an altered hyperbolic shape with a steeper initial curve than non-cooperative binding.
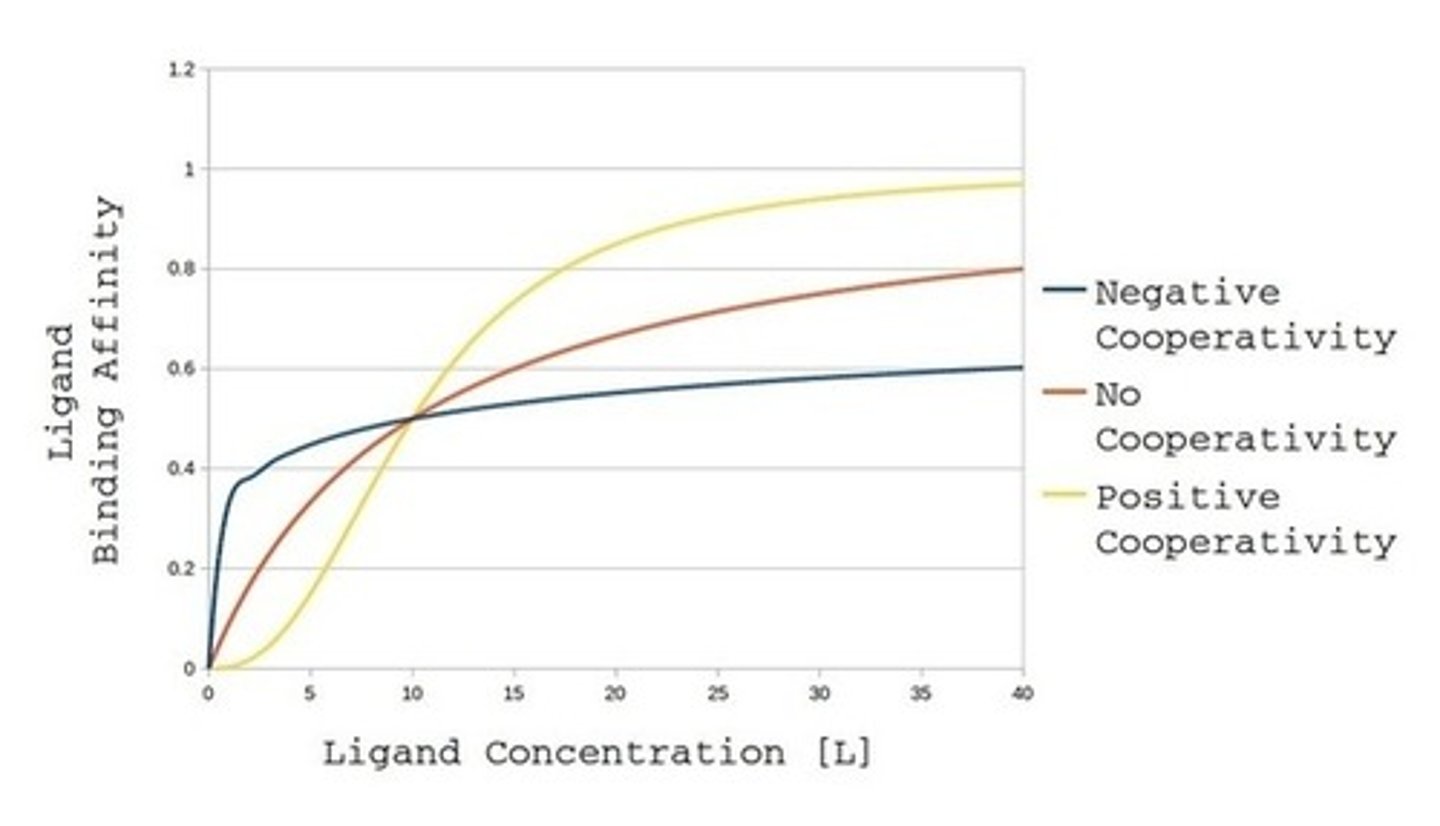
When talking about cooperativity, what can we infer about the enzyme?
I. The enzyme has multiple binding sites
II. The enzyme can bind to more than one substrate
III. The enzyme is immune to inhibitors
(A) I Only
(B) II Only
(C) I and II Only
(D) II and III Only
(C) I and II Only
When talking about cooperativity we can infer that the enzyme has multiple binding sites and can bind to more than one substrate.
Hemoglobin is written as Hb(O2)4. What type of cooperativity does hemoglobin have, and how does this affect oxygen binding?
(A) positive-cooperative binding
(B) negative-cooperative binding
(C) neutral-cooperative binding
(D) non-cooperative binding
(A) positive-cooperative binding
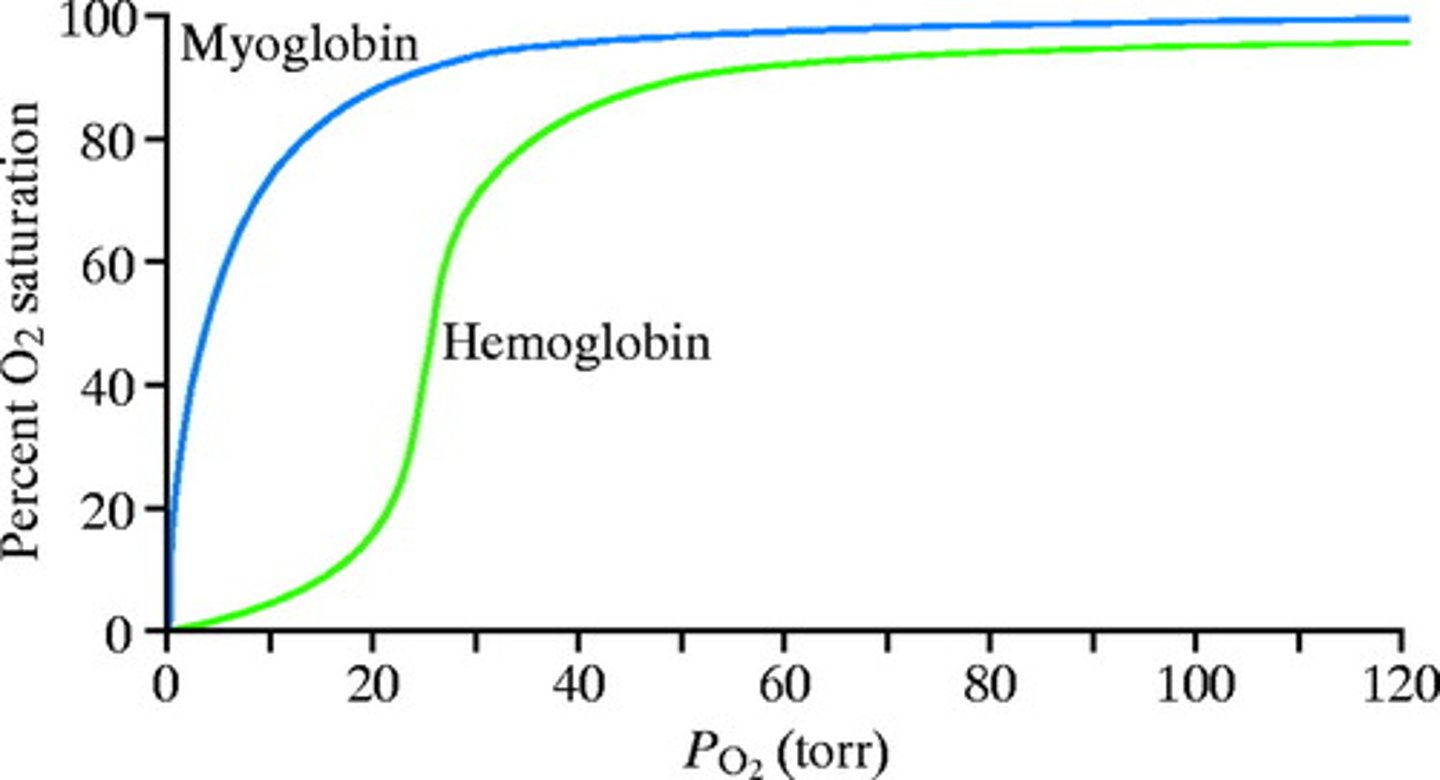
Hemoglobin exhibits positive cooperative binding. This means that after the first oxygen molecule binds, a second molecule of oxygen is more likely to bind because the affinity for that substrate increased. This affinity will continue increasing as more oxygen binds until the hemoglobin is saturated. Because of this, hemoglobin has a higher affinity for oxygen at higher partial pressures of oxygen.
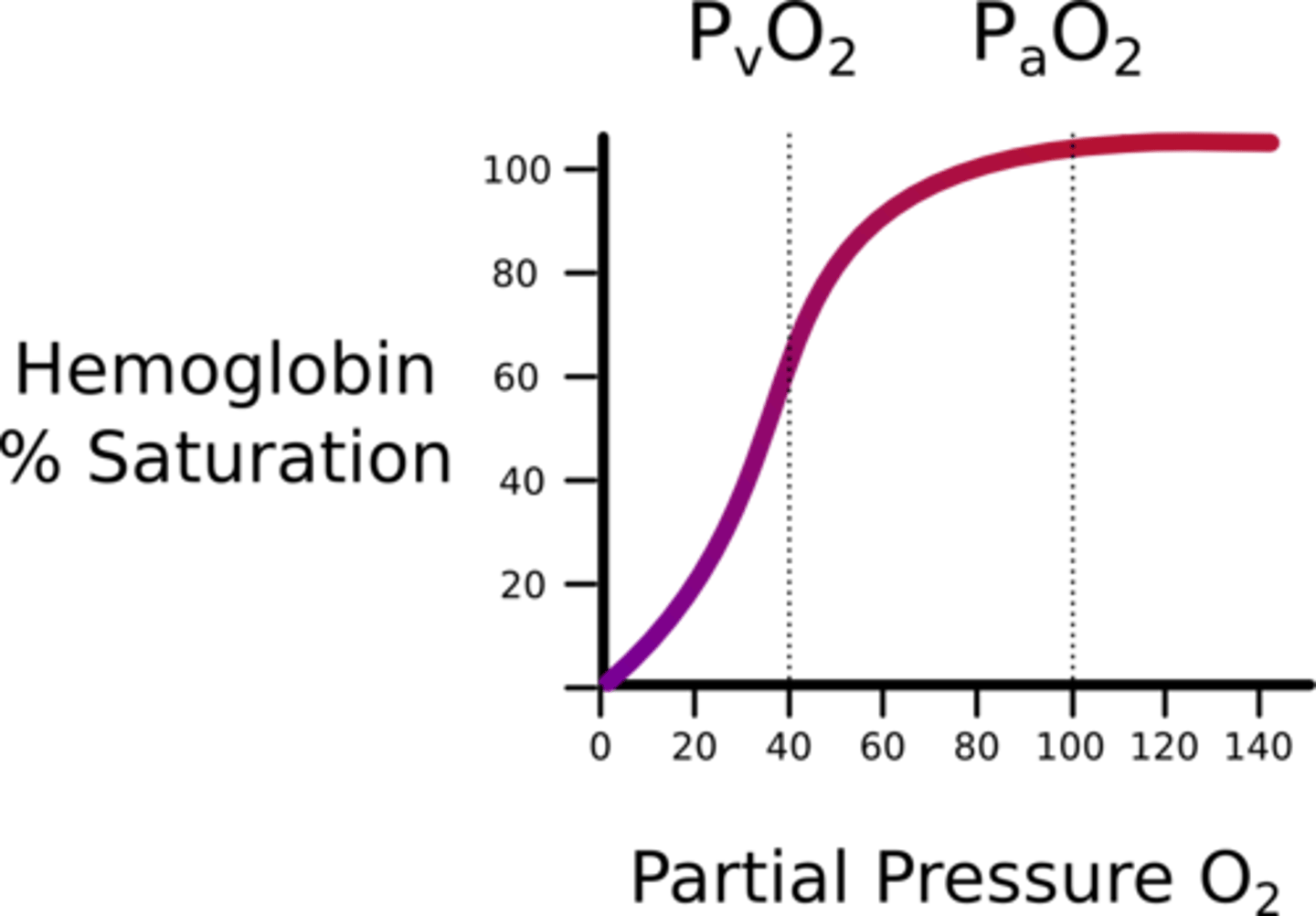
Myoglobin is written as Mb(O2). What type of cooperativity does myoglobin have and why?
(A) positive-cooperative binding
(B) negative-cooperative binding
(C) neutral-cooperative binding
(D) non-cooperative binding
(D) non-cooperative binding
Myoglobin exhibits non-cooperative binding because it can only bind one molecule of oxygen. In other words, there is no affinity for oxygen after the first molecule binds because there is no way to bind more!
Draw the percent saturation curves of Hemoglobin and Myoglobin. How do they differ?
Hemoglobin has a sigmoidal "S" shaped curve because it exhibits positive-cooperative binding. Myoglobin has a hyperbolic curve since it exhibits non-cooperative binding.
CRB Fill in the blanks: Binding Sites can exist in either the low-affinity ____________ state or the high-affinity _____________ state.
(A) R, S
(B) S, T
(C) T, S
(D) T, R
(D) T, R
Binding Sites can exist in either the low-affinity T (tense) state or the high-affinity R (relaxed) state.
CRB Which of the following statements about Positive Cooperativity are true?
I. Substrate binding to one binding site will encourage other binding sites to transition from the T state to the R state.
II. When substrates dissociate, the newly-empty binding site will almost instantaneously bind to another substrate to maintain that R-state.
III. When substrates dissociate, all subunits are more likely to transition from R state to the T state, promoting further dissociation.
(A) I only
(B) II only
(C) I and II only
(D) I and III only
(D) I and III only
Each of the following about Positive Cooperativity are true:
I. Substrate binding to one binding site will encourage other binding sites to transition from the T state to the R state.
III. When substrates dissociate, all subunits are more likely to transition from R state to the T state, promoting further dissociation.
CRB Fill in the blanks: Cooperativity is often compared using Hill's Coefficient. If Hill's coefficient is ________, then positive cooperative binding is occurring, and if Hill's coefficient is _________, then negative cooperative binding is occurring.
(A) > 0 , < 0
(B) < 0 , > 0
(C) < 1 , > 1
(D) > 1 , < 1
(D) > 1 , < 1
Cooperativity is often compared using Hill's Coefficient. If Hill's coefficient is > 1, then positive cooperative binding is occurring, and if Hill's coefficient is < 1, then negative cooperative binding is occurring.
CRB What type of binding is occuring if Hill's Coefficient is equal to 1?
If Hill's Coefficient is equal to 1, then the enzyme exhibits Non-Cooperative Binding.
Compare allosteric site vs. active site?
Allosteric sites are sites on the enzyme other than the active site to which a regulator can bind.
The active site is where the substrate binds.

CRB Which of the following is NOT going to affect all enzymes if it is drastically altered from physiological norms?
(A) Salinity
(B) Allosteric Inhibitor Concentrations
(C) Temperature
(D) pH
(B) Allosteric Inhibitor Concentrations
Allosteric Inhibitors do not affect all enzymes, only ones with allosteric sites compatible with that specific inhibitor.
Salinity, Temperature and pH could affect all enzymes if they are drastically altered.
CRB Drastically increasing which of the following could lead to denaturing an enzyme, decreasing its rate of reaction?
I. Temperature
II. pH
III. Salinity
(A) I only
(B) I and II only
(C) II and III only
(D) I, II and III
(D) I, II and III
Drastically increasing any of temperature, pH or Salinity could lead to enzyme denaturation.
CRB Compare how drastically increasing and decreasing pH can differently cause Denaturation.
Increasing pH will cause deprotonation of key residues in the active site and affect hydrogen bonding of the secondary structure. Decreasing pH could protonate key residues in the active site, also affecting the hydrogen bonding of the secondary structure, and could also protonate the Cystines, breaking all disulfide bonds.
CRB How could dramatically increasing the Salinity of an solution with enzymes denature the enzyme?
(A) It could disrupt the Primary Structure by breaking only covalent bonds.
(B) It could disrupt Secondary Structure by disrupting only Hydrogen Bonds.
(C) It could disrupt Tertiary and Quaternary Structure by disrupting only Ionic Bonds.
(D) It could disrupt Secondary, Tertiary and Quaternary Structure by disrupting both Hydrogen and Ionic Bonds.
(D) Increased Salinity could disrupt Secondary, Tertiary and Quaternary Structure by disrupting both Hydrogen and Ionic Bonds.
Compare the two types of enzyme regulators -- allosteric activators vs. inhibitors. How does each affect Km and Vmax?
Allosteric activators increase enzyme activity. They increase Vmax and decrease Km.
Allosteric inhibitors decrease enzyme activity. They decrease Vmax and increase Km.
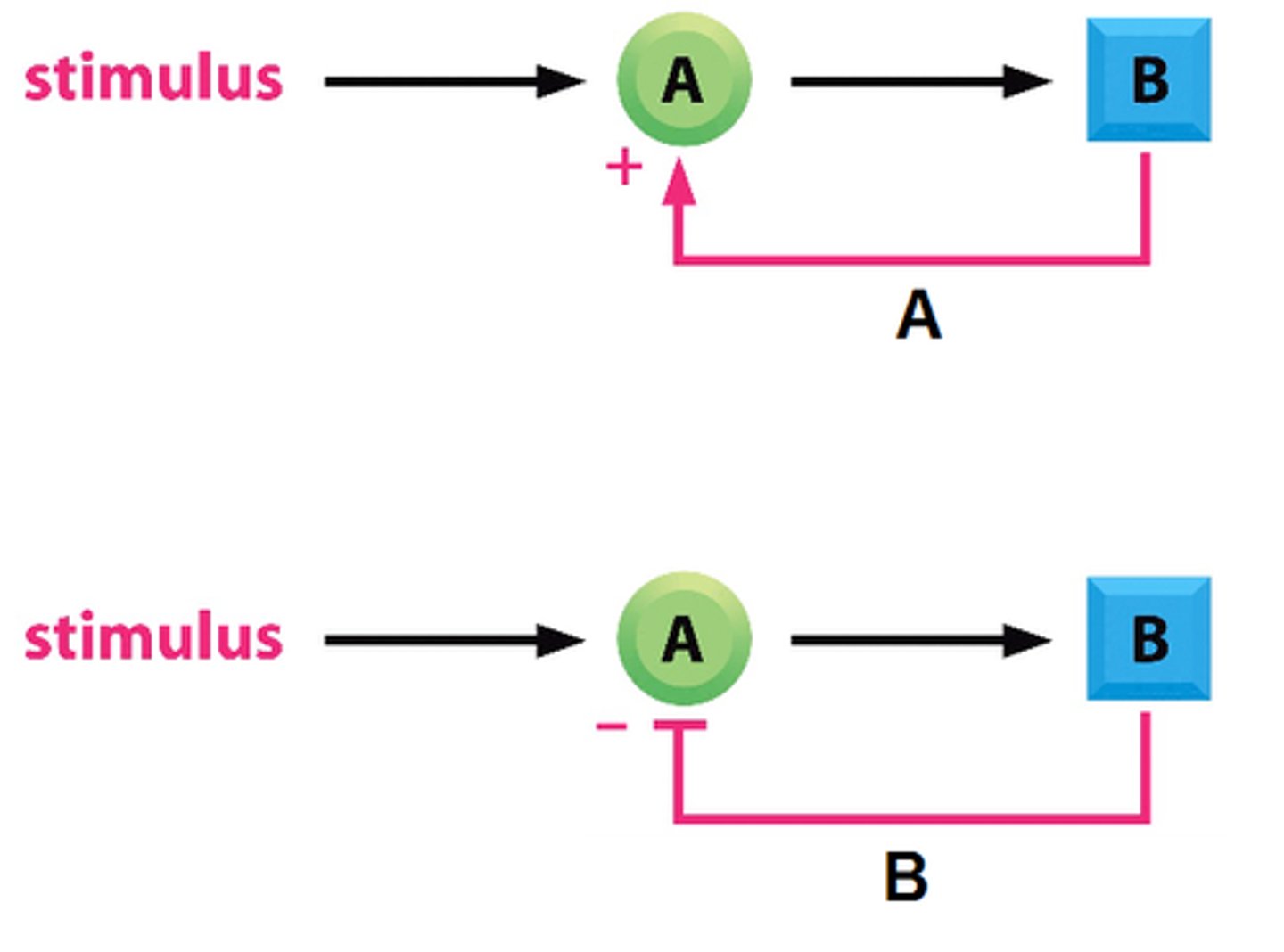
What is the difference/similarity between a positive feedback loop versus a negative feedback loop?
Both a positive feedback loop and a negative feedback loop consist of a downstream product that regulates upstream reactions. The difference is that the output/product of positive feedback loop acts as an activator and amplifies the system, whereas the output/product of a negative feedback loop inhibits the system.
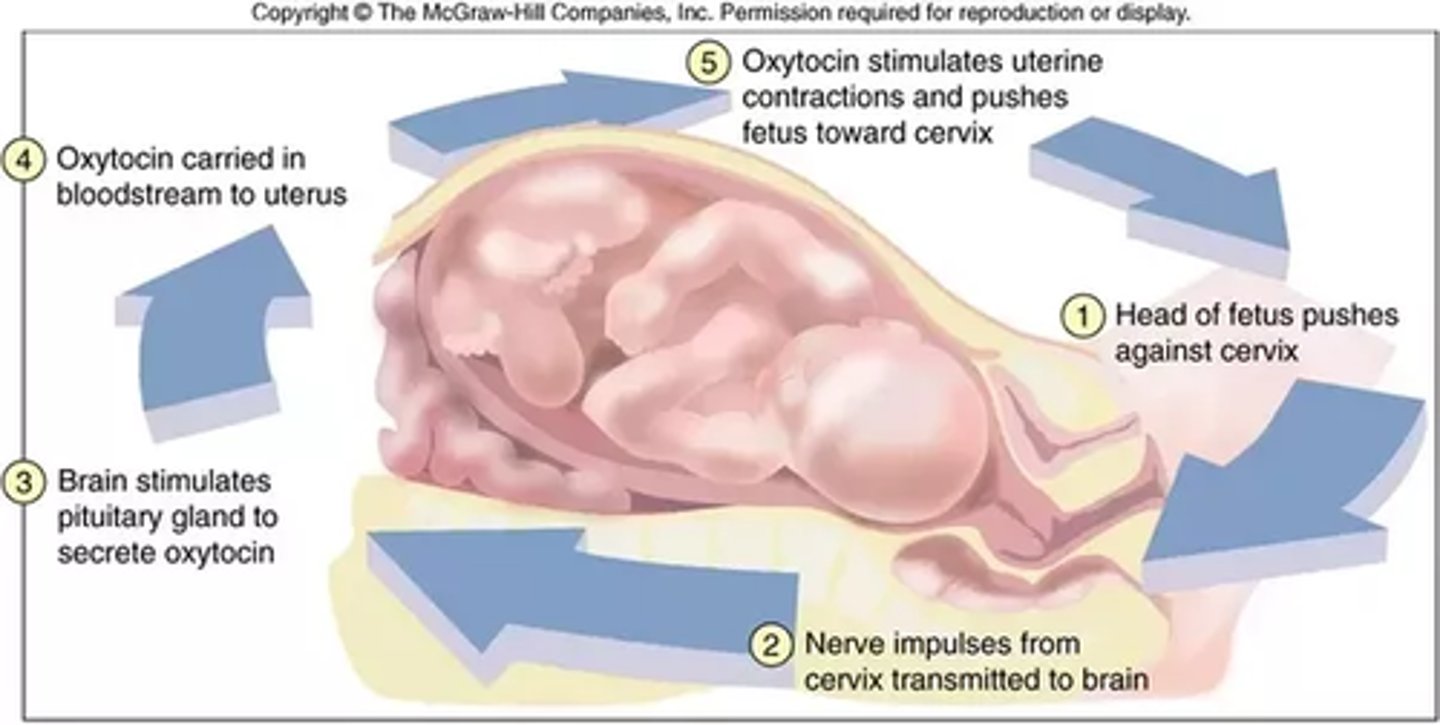
During labor, the baby's head is pushed downward and results in increased pressure on the cervix. This contraction stimulates receptor cells which send a chemical signal to the brain, causing an increased release of oxytocin. The release of oxytocin stimulates further contractions. These contractions stimulate further oxytocin release until the baby is born. What kind of feedback loop is this? Positive or Negative?
This feedback loop is an example of a positive feedback loop because oxytocin acts as an activator, amplifying the system to perform more contractions.
Blood pressure must be high enough to pump blood throughout the body, but not high enough to cause damage. While the heart is pumping, baroreceptors detect the pressure of the blood going through arteries. If the pressure is too high, the baroreceptors will send an electrochemical signal to the brain via the glossopharyngeal nerve. The brain then sends a chemical signal to the heart to decrease the pressure. Is this an example of a positive or negative feedback loop?
This is an example of a negative feedback loop because the output (chemical signal to decrease blood pressure) inhibits the input (high blood pressure detected by baroreceptors).
What is a homotropic inhibitor/activator versus a heterotropic inhibitor/activator?
A homotropic inhibitor/activator is a molecule that acts as both a substrate and an inhibitor/activator.
A heterotropic inhibitor/activator is strictly a regulating molecule and not an active site substrate.
During glycolysis, phosphofructokinase catalyzes the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate using ATP. However, high levels of ATP inhibit phosphofructokinase. In this scenario, ATP is acting as a:
(A) Homotropic inhibitor
(B) Homotropic activator
(C) Heterotropic inhibitor
(D) Heterotropic activator
(A) Homotropic inhibitor
ATP is acting as a homotropic inhibitor because it acts as a substrate for the enzyme and as an inhibitor.
Also, because the goal of glycolysis is to produce ATP, this can also be considered a negative feedback loop (output ATP reduces the input of phosphorylating fructose to start glycolysis).
During glycolysis, phosphofructokinase catalyzes the conversion of fructose to fructose-6-bisphosphate using ATP. High levels of AMP enhance the activity of phosphofructokinase. In this scenario, AMP is acting as a:
(A) Homotropic inhibitor
(B) Homotropic activator
(C) Heterotropic inhibitor
(D) Heterotropic activator
(D) Heterotropic activator
AMP is acting as a heterotropic activator, because it does not act as a substrate for the enzyme that it activates.
A single reaction in a pathway would be a great control point for regulation if it has a very:
(A) positive ΔG.
(B) negative ΔG.
(C) positive ΔS.
(D) negative ΔS.
(B) negative ΔG.
A reaction with a negative ΔG would be a great "committing" step for a pathway because that reaction is unlikely to be reversed. That reaction's products would then be committed to moving forward in the pathway. Phosphofructokinase's reaction has a very negative ΔG; thus, it is a great control point for glycolysis.
CRB There are a variety of ways to separate proteins based on their different characteristics. Which of the following would you NOT expect to be used to separate proteins?
(A) Electrophoresis
(B) Isoelectric Focusing
(C) X-ray Crystallography
(D) Chromatography
(C) X-ray Crystallography
X-ray Crystallography could be used once a protein has been purified to learn about its structure.
Electrophoresis, Isoelectric Focusing and Chromatography are all techniques used to separate proteins.
(Note that there is a whole lesson on Chromatography in Chemistry II, so we will not discuss it further here).
CRB Match each of the following types of Gel Electrophoresis with their descriptions.
I. Polyacrylamide Gel.
II. Native PAGE
III. SDS-PAGE
(A) Can only give mass-to-charge ratios, so could have decreased separation of proteins with different masses or charges. This does NOT denature the protein.
(B) The typical medium used for protein electrophoresis.
(C) Uses a Detergent to break all noncovalent interactions, including affecting the charges of proteins. They are only separated based on size.
Note that PAGE stands for Polyacrylamide Gel Electrophoresis
I. Polyacrylamide Gel - (B) The typical medium used for protein electrophoresis.
II. Native PAGE - (A) Can only give mass-to-charge ratios, so could have decreased separation of proteins with different masses or charges. This does NOT denature the protein.
III. SDS-PAGE - (C) Uses a Detergent to break all noncovalent interactions, including affecting the charges of proteins. They are only separated based on size.
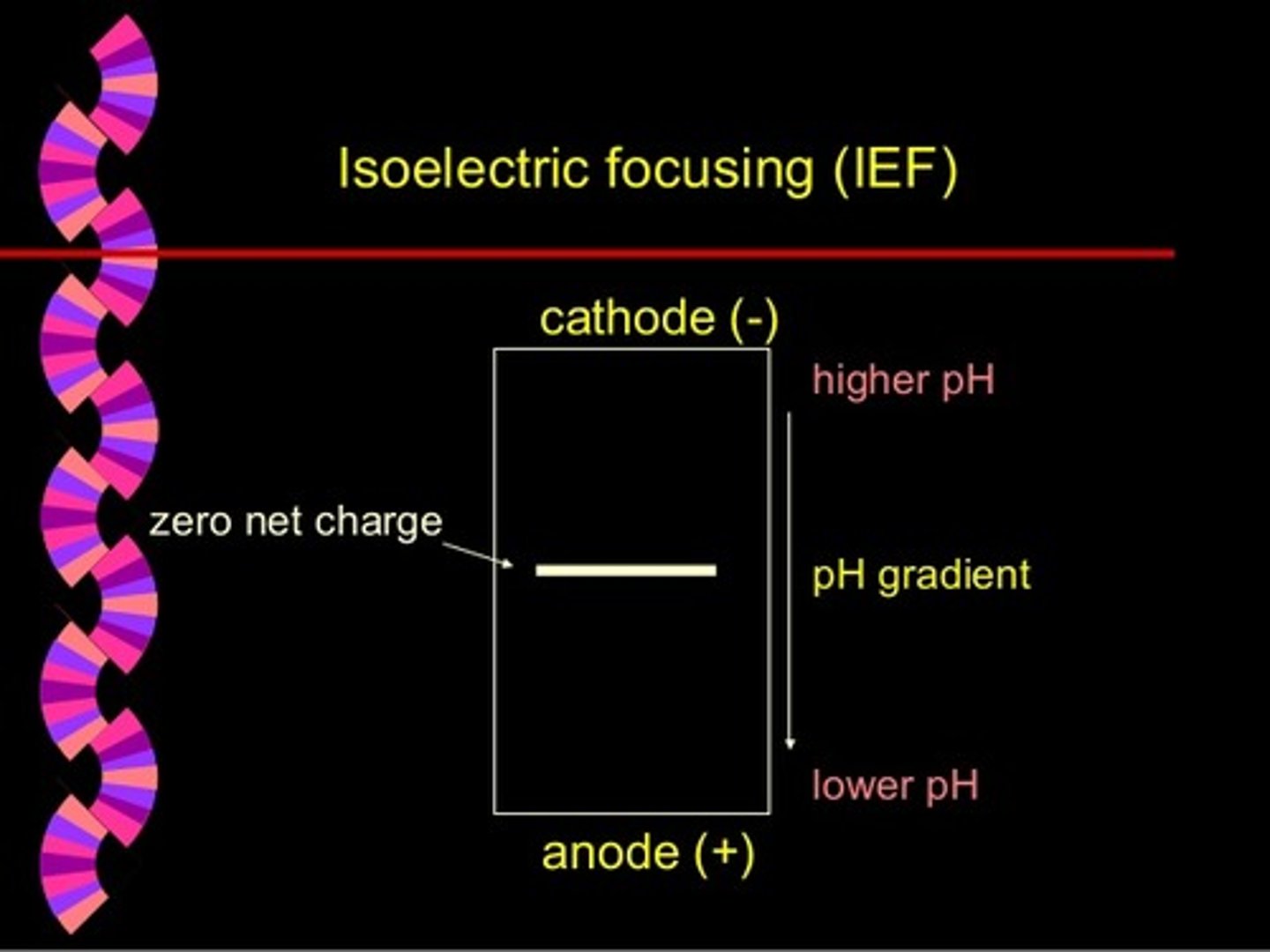
CRB Which of the following descriptions of Isoelectric Focusing are true?
I. The gel that the samples are separated in has a pH gradient, ranging from acidic to basic.
II. The protein will settle at the pH where the protein's R-groups are fully deprotonated.
III. As proteins move towards the Cathode, the pH increases.
(A) I only
(B) I and III only
(C) II and III only
(D) I, II and III
(B) I and III only
Each of the following statements are true about Isoelectric Focusing:
I. The gel that the samples are separated in has a pH gradient, ranging from acidic to basic.
II. The protein will settle at the pH equal to the protein's pI.
III. As proteins move towards the Cathode, the pH increases.
CRB True or false? In isoelectric focusing, the proteins are loaded near the Anode, where there is a low pH and the proteins will be protonated and positively charged.
True. In isoelectric focusing, the proteins are loaded near the Anode, where there is a low pH and the proteins will be protonated and positively charged.
CRB One of the ways that Proteins are analyzed are through Amino Acid Composition. Which of the following is a sequential digestion technique that can sequence 50-70 amino acids from the N-terminus?
(A) NMR
(B) Bradford Assay
(C) Edman Degradation
(D) Lowry Reagent Assay
(C) Edman Degradation
Edman Degradation is a sequential digestion technique that can sequence 50-70 amino acids from the N-terminus, giving us knowledge about a protein's Primary Structure.
CRB True or false? UV Spectroscopy is typically used to determine the concentration of proteins in a sample because it requires little decontamination.
False. UV Spectroscopy is typically used to determine the concentration of proteins in a sample, but it is very sensitive to contaminants in the sample!
CRB The Bradford Protein Assay is one of the most common and simplest Protein Assays. Which of the following is NOT a true statement about Bradford Protein Assays?
(A) It mixes dissolved proteins with a green-brown dye that will turn blue.
(B) The dye accepts electrons from the amino acid groups, triggering that color change.
(C) Ionic interactions stabilize the blue form of this dye, and then Spectrophotometry can be used.
(D) Samples must be compared to an appropriate Standard Curve to determine the concentration of the protein.
(B) The dye accepts electrons from the amino acid groups, triggering that color change.
The Dye actually Donates protons to the Amino Acids.
Also note that Bradford Protein Assays are sensitive to having multiple proteins mixed in one sample and to having too dilute a sample or detergent in the sample.