Chapter 10 End of Chapter Questions
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
Knowledge and Comprehension
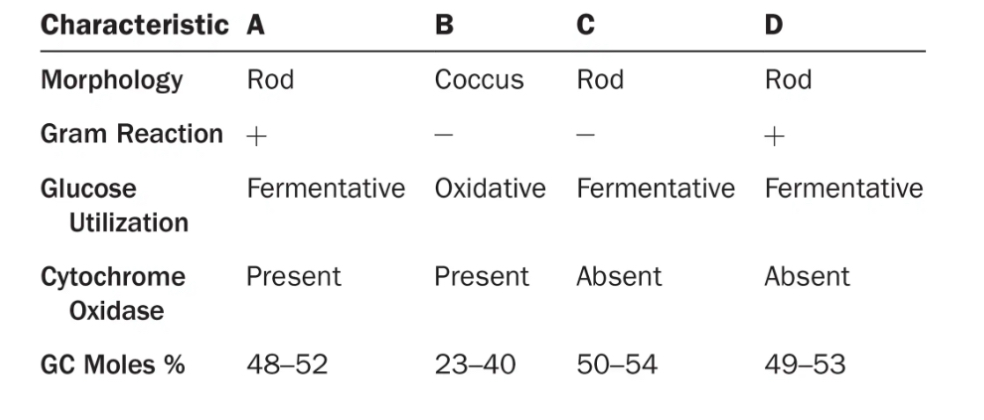
Which of the following organisms are most closely related?
Are any two the same species?
On what did you base your answer?
A and D appear to be most closely related because they have similar GC moles %.
No, they all have differences from one another as shown in the graph
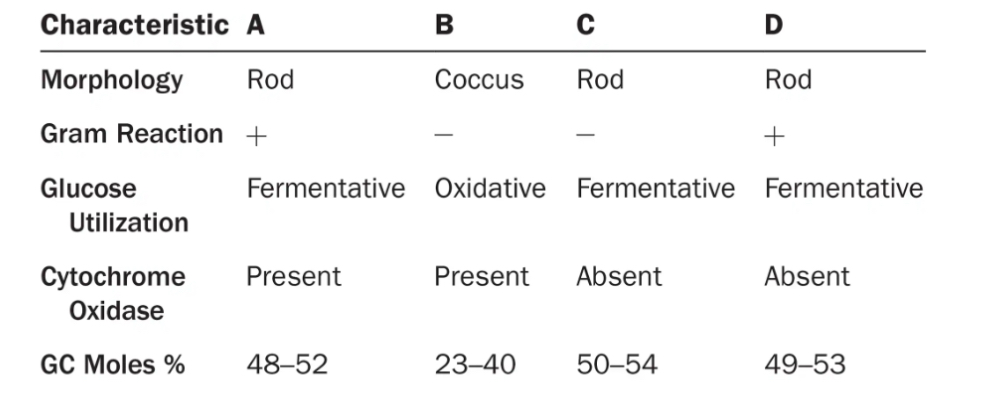

Which of these organisms are most closely related? Compare this answer with your response to review question 1.
A and D are most closely related which follows up with our observations from question 1
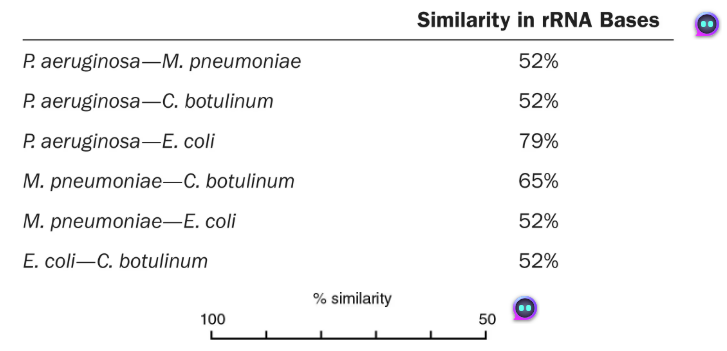
DRAW IT Use the following rRNA information to construct a cladogram for some of the organisms used in question 4.
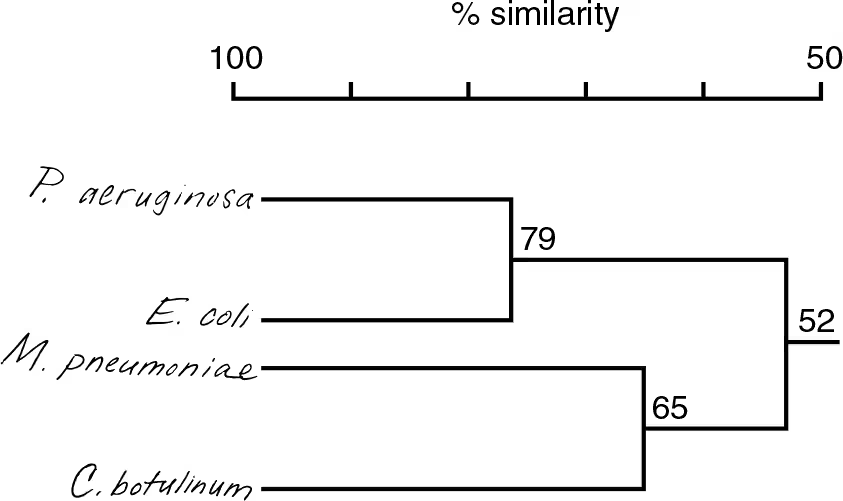
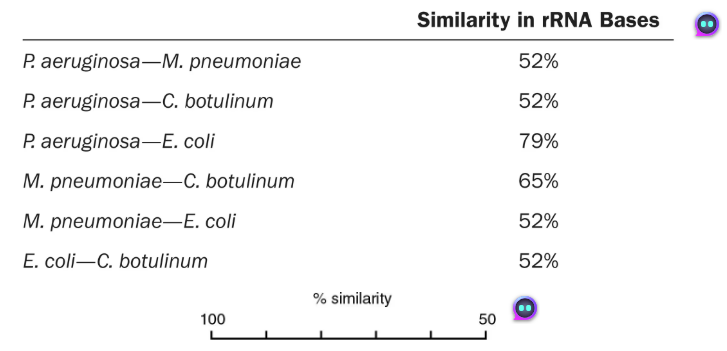
What is the purpose of a cladogram?
Shows degree of relatedness between organisms
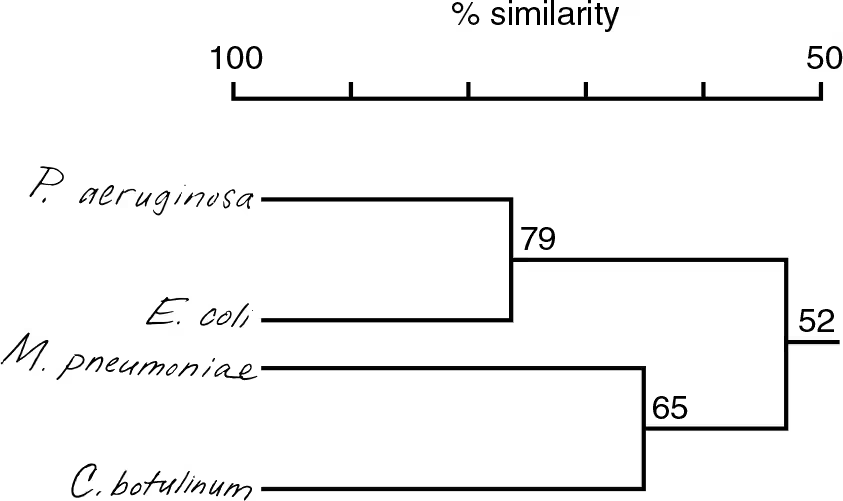
How does your cladogram differ from a dichotomous key for these organisms?
A dichotomous key can be used for identification but doesn’t show relatedness like the cladogram. Mycoplasma and Escherichia are on one branch in the key, but the cladogram indicates Mycoplasma is more closely related to Clostridium.
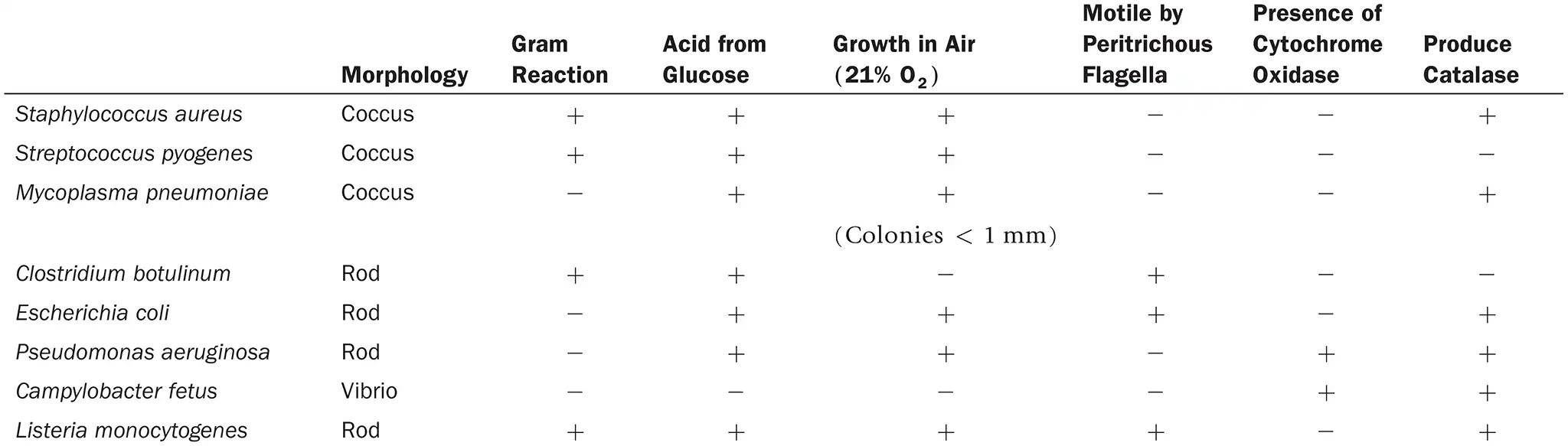
DRAW IT Use the information in the following table to complete the dichotomous key to these organisms.
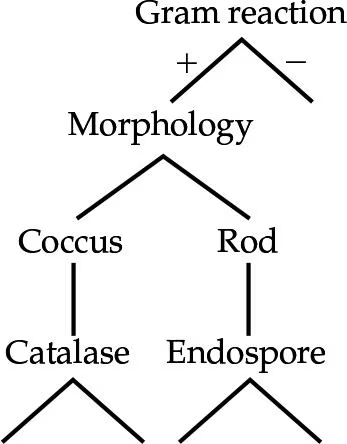
(view image) One possible key is shown. Alternative keys could be made starting with morphology or glucose fermentation
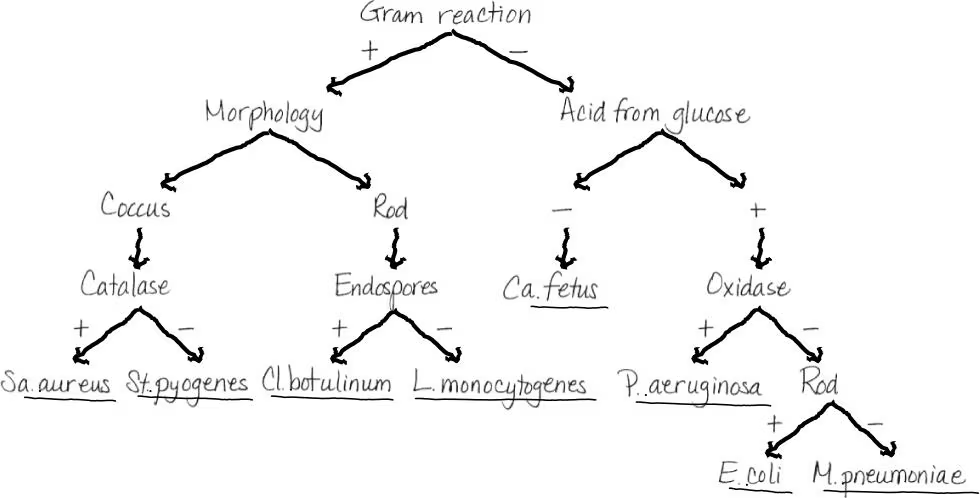
What is the purpose of a dichotomous key?
Helps identify organisms/objects through a series of statements/questions each with 2 possible outcomes and ultimately guiding the user to the correct identification. (example shown)

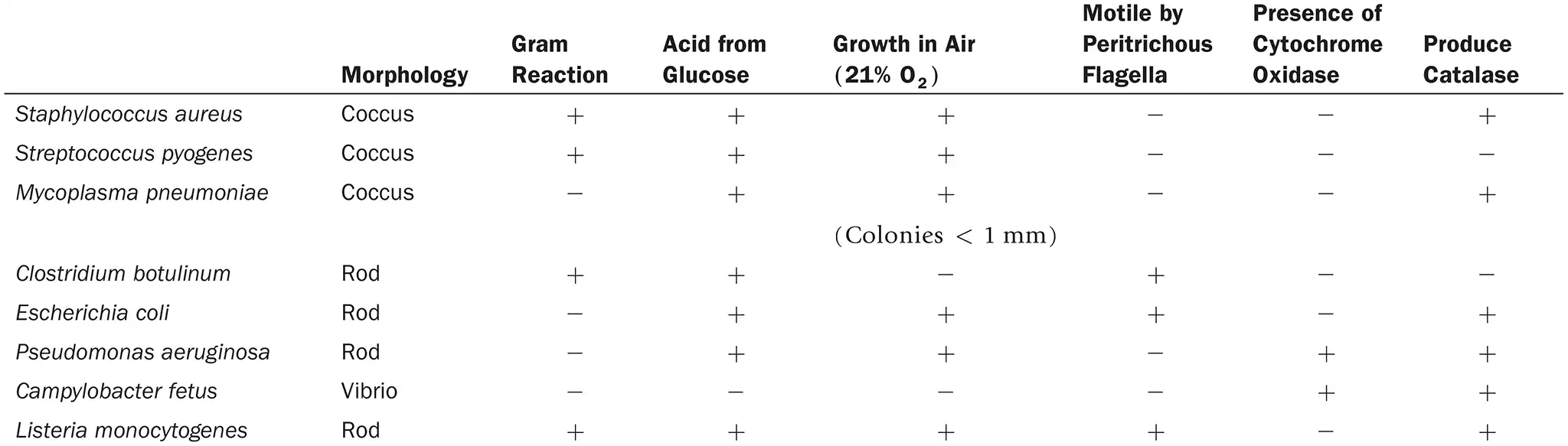
Look up each genus in Chapter 11, and provide an example of why this organism is of interest to humans.
Staphylococcus aureus:
Streptococcus pyogenes:
Mycoplasma pneumoniae:
Clostridium botulinum:
Escherichia coli:
Pseudomonas aeruginosa:
Campylobacter fetus:
Listeria monocytogenes:
answers are from some light research, didn’t use the textbook
Staphylococcus aureus: Causes skin infections, pneumonia, endocarditis, and sepsis, MRSA
Streptococcus pyogenes: Causes strep throat, scarlet fever, impetigo, and necrotizing fasciitis
Mycoplasma pneumoniae: causes pneumonia
Clostridium botulinum: produces botulinum toxin —> causes botulism
Escherichia coli: part of normal gut flora, pathogenic strains cause foodborne illness, bloody diarrhea, and kidney damage
Pseudomonas aeruginosa: Causes opportunistic infections, especially in burn victims, cancer patients, and people with cystic fibrosis, Highly antibiotic-resistant, forming biofilms that protect it, A major concern in hospital-acquired infections (HAIs).
Campylobacter fetus: Linked to gastroenteritis, bacteremia, and reproductive issues
Listeria monocytogenes: listeria causes listeriosis
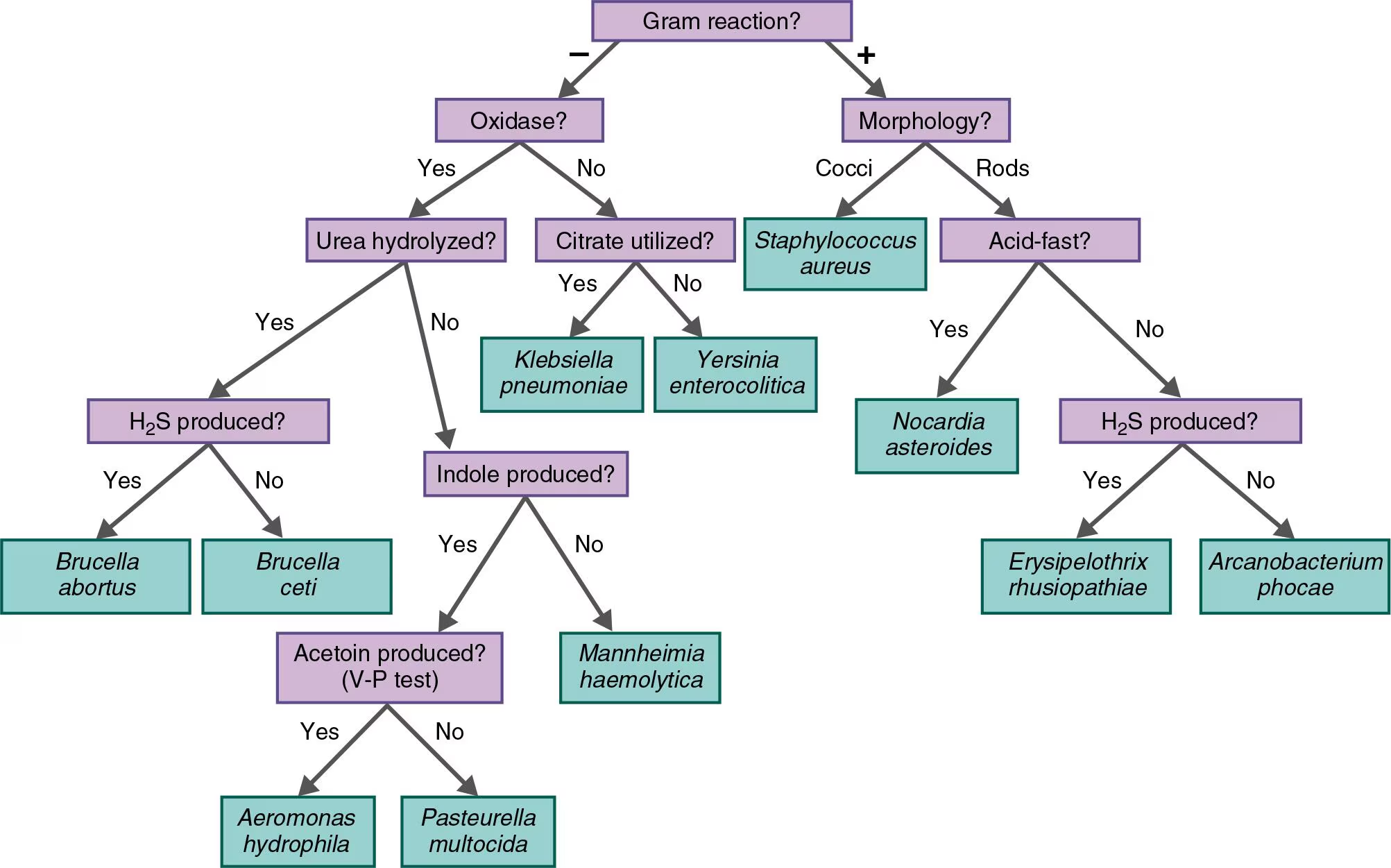
NAME IT Use the key in the Clinical Focus box to identify the gram-negative, oxidase-positive rod causing pneumonia in a sea otter. It is H2S-positive, indole-negative, and urease-positive.
Brucella abortus
Multiple Choice
Bergey’s Manual of Systematics of Archaea and Bacteria differs from Bergey’s Manual of Determinative Bacteriology in that the former
groups bacteria into species.
groups bacteria according to phylogenetic relationships.
groups bacteria according to pathogenic properties.
groups bacteria into 19 species.
all of the above
2
Bacillus and Lactobacillus are not in the same order. This indicates that which one of the following is not sufficient to assign an organism to a taxon?
biochemical characteristics
amino acid sequencing
phage typing
serology
morphological characteristics
5
Which of the following is used to classify organisms into the Kingdom Fungi?
ability to photosynthesize; possess a cell wall
unicellular; possess cell wall; prokaryotic
unicellular; lacking cell wall; eukaryotic
absorptive; possess cell wall; eukaryotic
ingestive; lacking cell wall; multicellular; prokaryotic
4
Which of the following is false about scientific nomenclature?
Each name is specific.
Names vary with geographical location.
The names are standardized.
Each name consists of a genus and specific epithet.
It was first designed by Linnaeus.
2
You could identify an unknown bacterium by all of the following except
hybridizing a DNA probe from a known bacterium with the unknown’s DNA.
making a fatty acid profile of the unknown.
specific antiserum agglutinating the unknown.
ribosomal RNA sequencing.
percentage of guanine and cytosine
5
The wall-less mycoplasmas are considered to be related to gram-positive bacteria. Which of the following would provide the most compelling evidence for this?
They share common rRNA sequences.
Some gram-positive bacteria and some mycoplasmas produce catalase.
Both groups are prokaryotic.
Some gram-positive bacteria and some mycoplasmas have coccus-shaped cells.
Both groups contain human pathogens.
1
Into which group would you place a multicellular organism that has a mouth and lives inside the human liver?
Animalia
Fungi
Plantae
Bacillota (gram-positive bacteria)
Pseudomonadota (gram-negative bacteria)
1
Into which group would you place a photosynthetic organism that lacks a nucleus and has a thin peptidoglycan wall surrounded by an outer membrane?
Animalia
Fungi
Plantae
Bacillota (gram-positive bacteria)
Pseudomonadota (gram-negative bacteria)
5

Which is (are) found in all three domains?
2,6
5
2,4,6
1,3,5
all six
1

Which is (are) found only in prokaryotes?
1,4,6
3,5
1,2
4
2,4,5
2
Analysis
The GC content of Micrococcus is 66–75 moles %, and of Staphylococcus, 30–40 moles %. According to this information, would you conclude that these two genera are closely related?
No, the 2 species aren’t related because their G-C content values are very far apart (difference of >10%)
66-30= 36 and 75- 40= 35% = 35-36% > 10% so - not related
(G-C + A-T = 100%)
Describe the use of a DNA probe and PCR for:
rapid identification of an unknown bacterium.
DNA probe: give us library of known bacteria to compare unknown bacteria to on plate
PCR: allows for replication and isolation of one strand of DNA to match to DNA probe
Describe the use of a DNA probe and PCR for:
determining which of a group of bacteria are most closely related.
DNA probe: allows us to match enzymes from sample to determine molecular %
PCR: allows for replication and isolation of one strand of DNA to match to DNA probe
SF medium is a selective medium, developed in the 1940s, to test for fecal contamination of milk and water. Only certain gram-positive cocci can grow in this medium.
Why is it named SF?
It is named for its target organism, Streptococcus Faecalis.
SF medium is a selective medium, developed in the 1940s, to test for fecal contamination of milk and water. Only certain gram-positive cocci can grow in this medium.
Using this medium, which genus will you culture? (Hint: Refer to the discussion of Scientific Nomenclature.)
Streptococcus genus
Clinical Applications and Evaluation
A 55-year-old veterinarian was admitted to a hospital with a 2-day history of fever, chest pain, and cough. Gram-positive cocci were detected in his sputum, and he was treated for lobar pneumonia with penicillin. The next day, another Gram stain of his sputum revealed gram-negative rods, and he was switched to ampicillin and gentamicin. A sputum culture showed biochemically inactive gram-negative rods identified as Pantoea (Enterobacter) agglomerans. After fluorescent-antibody staining and phage typing, Y. pestis was identified in the patient’s sputum and blood, and chloramphenicol and tetracycline were administered. The patient died 3 days after admission to the hospital. Tetracycline was given to his 220 contacts (hospital personnel, family, and co-workers).
What disease did the patient have?
Black Death/Plague
A 55-year-old veterinarian was admitted to a hospital with a 2-day history of fever, chest pain, and cough. Gram-positive cocci were detected in his sputum, and he was treated for lobar pneumonia with penicillin. The next day, another Gram stain of his sputum revealed gram-negative rods, and he was switched to ampicillin and gentamicin. A sputum culture showed biochemically inactive gram-negative rods identified as Pantoea (Enterobacter) agglomerans. After fluorescent-antibody staining and phage typing, Y. pestis was identified in the patient’s sputum and blood, and chloramphenicol and tetracycline were administered. The patient died 3 days after admission to the hospital. Tetracycline was given to his 220 contacts (hospital personnel, family, and co-workers).
Discuss what went wrong in the diagnosis and how his death might have been prevented.
His sputum culture should have underwent more testing prior to antibiotic administration.
A 55-year-old veterinarian was admitted to a hospital with a 2-day history of fever, chest pain, and cough. Gram-positive cocci were detected in his sputum, and he was treated for lobar pneumonia with penicillin. The next day, another Gram stain of his sputum revealed gram-negative rods, and he was switched to ampicillin and gentamicin. A sputum culture showed biochemically inactive gram-negative rods identified as Pantoea (Enterobacter) agglomerans. After fluorescent-antibody staining and phage typing, Y. pestis was identified in the patient’s sputum and blood, and chloramphenicol and tetracycline were administered. The patient died 3 days after admission to the hospital. Tetracycline was given to his 220 contacts (hospital personnel, family, and co-workers).
Why were the 220 other people treated? (Hint: Refer to Chapter 23.)
Black Death/Plague is highly virulent

A 6-year-old girl was admitted to a hospital with endocarditis. Blood cultures showed a gram-positive, aerobic rod identified by the hospital laboratory as Corynebacterium xerosis. The girl died after 6 weeks of treatment with intravenous penicillin and chloramphenicol. The bacterium was tested by another laboratory and identified as C. diphtheriae. The following test results were obtained by each laboratory: (view image)
Provide a possible explanation for the incorrect identification.
The discrepancy in the sucrose fermentation results between the labs, and the lack of the toxin production test in the hospital lab, which turned out positive in the other lab

A 6-year-old girl was admitted to a hospital with endocarditis. Blood cultures showed a gram-positive, aerobic rod identified by the hospital laboratory as Corynebacterium xerosis. The girl died after 6 weeks of treatment with intravenous penicillin and chloramphenicol. The bacterium was tested by another laboratory and identified as C. diphtheriae. The following test results were obtained by each laboratory: (view image)
What are the potential public health consequences of misidentifying C. diphtheriae? (Hint: Refer to Chapter 24.)
Diphtheria outbreak, diptheria causes respiratory distress and heart failure to some. It is commonly associated with kids and teens.
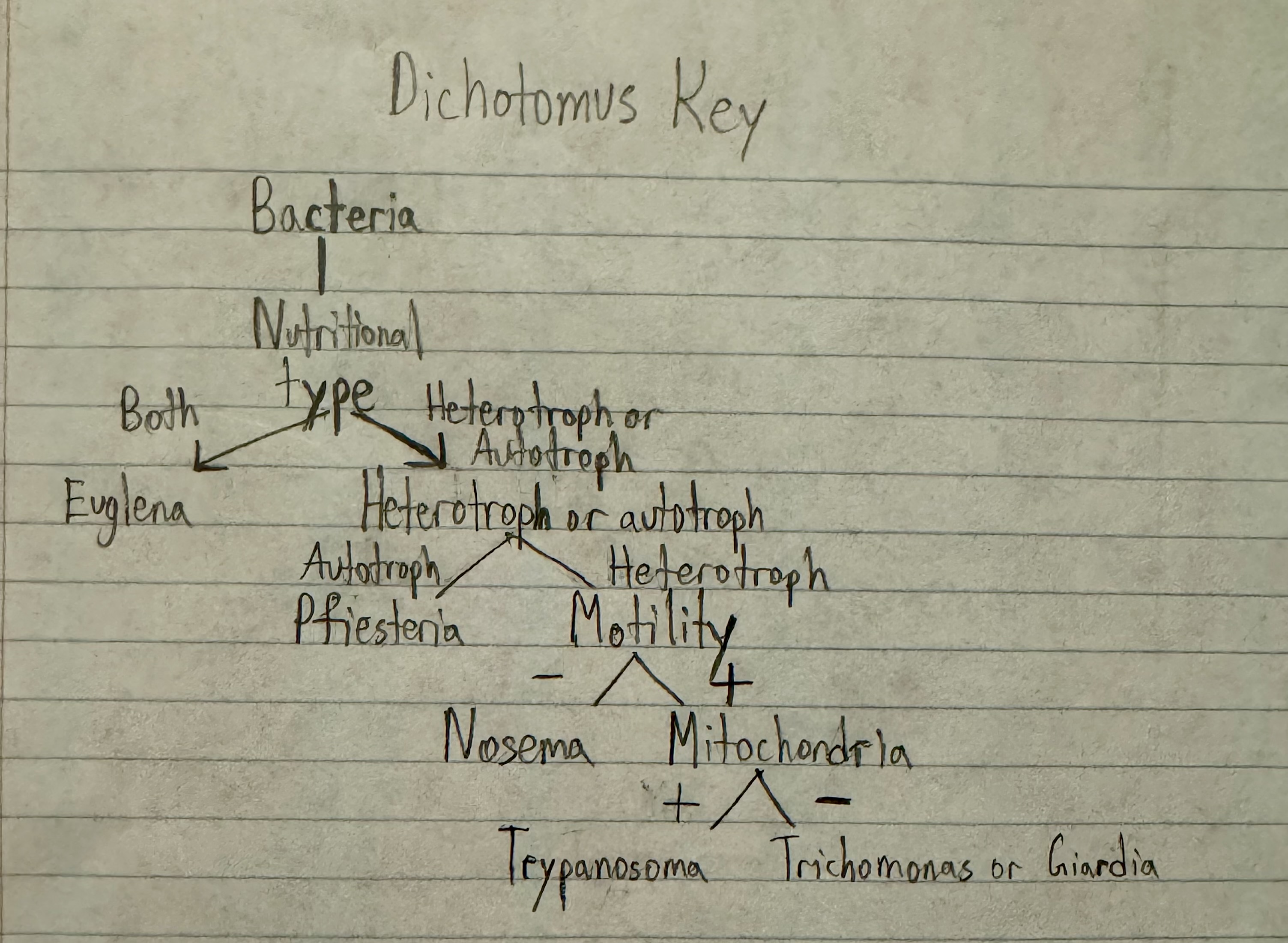
Using the following information, create a dichotomous key for distinguishing these unicellular organisms.
Which cause human disease?
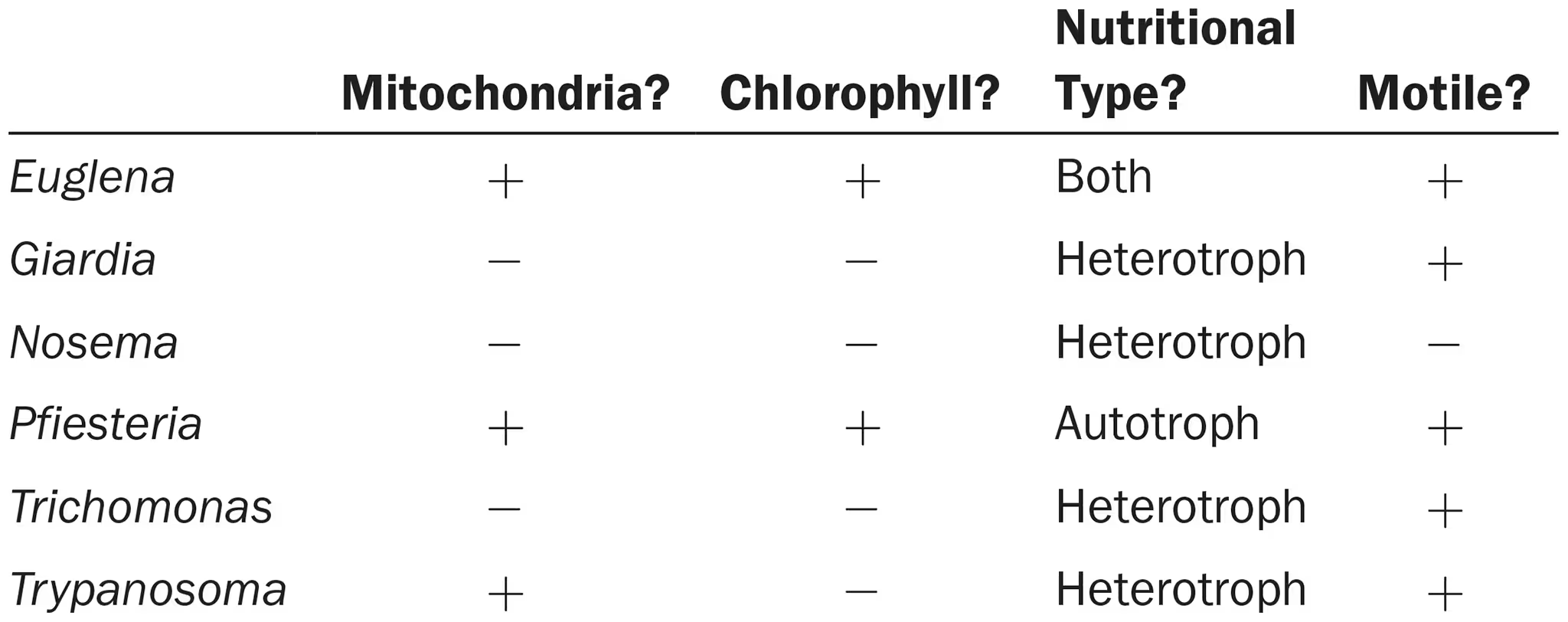
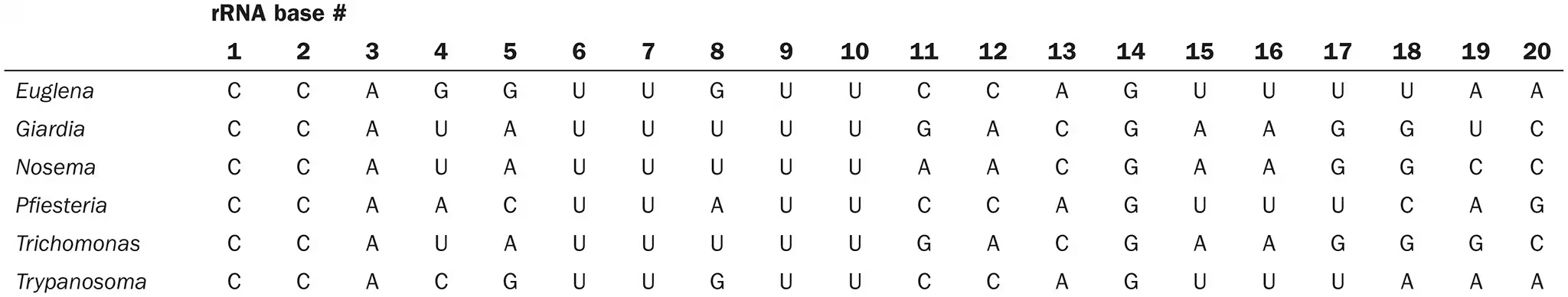
Using the additional information shown next, create a cladogram for these organisms. Do your two keys differ? Explain why. Which key is more useful for laboratory identification? For classification?
view image
euglena? (some species secrete toxins harmful to humans), giardia, pfiesteria, trichomonas, trypanosoma
idk, yes because the cladogram shows degree of relatedness while the dichotomus key doesn’t, dichotomus key more useful for identification? and cladogram more useful for classification?