Protein Structure and Function
1/62
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
63 Terms
Define Amino acids
monomeric building blocks of proteins. Small organic molecule containing an amino group and a carboxyl group.
Define peptide
a short chain of amino acids linked by peptide bonds.
Define polypeptide
A longer chain of amino acids joined via peptide bonds.
Define primary structure
The full amino acid sequence of a protein, where amino acids are linked together by covalent peptide bonds to form the polypeptide chain.
Define secondary structure
The local interactions between stretches of a polypeptide chain, forming structures including α-helices and β-pleated sheets.
• α-Helix: A coiled structure stabilized by hydrogen bonds.
• β-Sheet: A sheet-like structure formed by hydrogen bonds between strands.
• Turns/Loops: Non-regular structures that connect secondary structural elements.
Define tertiary structure
Folding of the α-helices and β-pleated sheets into a 3D structure
Define quaternary structure
A protein structure consisting of several tertiary structure subunits.
Define hydrogen bonds
Bonds between polar groups, critical for stabilizing secondary, tertiary, and quaternary structures
Define disulfide bonds
Covalent bonds between cysteine residues that stabilize the protein’s tertiary or quaternary structure
Define electrostatic interactions
Interactions between charged amino acid side chains (e.g., lysine, glutamate)
Define van de waals forces
Weak attractions between atoms in close proximity, contributing to
protein stability
Define molecular chaperone
proteins that assist during protein folding, preventing misfolding and the formation of aggregates.
Define native state
The fully folded, functional form of a protein
Define protein denaturation
the disruption and possible destruction of both the secondary and
tertiary structures. Depending on the type of disruption it can be reversible (chemical disruption) or irreversible (heat).
Define protein aggregation
Clustering of misfolded proteins, often associated with the pathology of diseases such as Alzheimer’s Disease.
Define protein domain
a conserved part of a given protein sequence and tertiary structure that can function and exist independently of the rest of the protein chain. Usually, they are responsible for a particular function or interaction
Define entropy
is a concept from thermodynamics that measures the degree of disorder or randomness in a system. In relation to protein folding, it is a measure of the number of possible ways to arrange the protein
6 functions of proteins
Structural proteins
Secreted structural proteins
Molecular motors
Intracellular proteins
Secreted proteins
Membrane bound proteins
Describe structural proteins
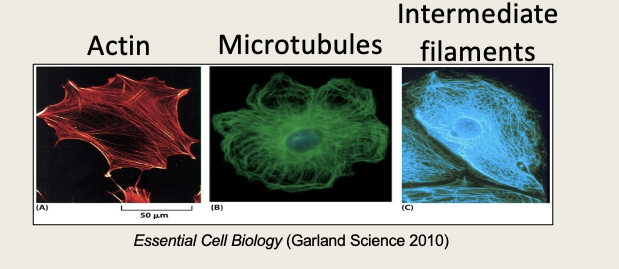
Define intracellular proteins
Cytosolic Signalling molecules- transfer signals from the plasma membrane to the nucleus
Example of secreted structural proteins
Collagen- cartilage
Example of molecular motors
DNA helicase – opens the DNA double helix during DNA synthesis\
Examples of membrane bound proteins
Receptors – recognition sites for specific molecules that transmit signals from outside to inside the cell
Mechanisms of protein variation
Single nucleotide polymorphisms (SNPs)
Alternative splicing
Post-translational modifications
Amino acid structure
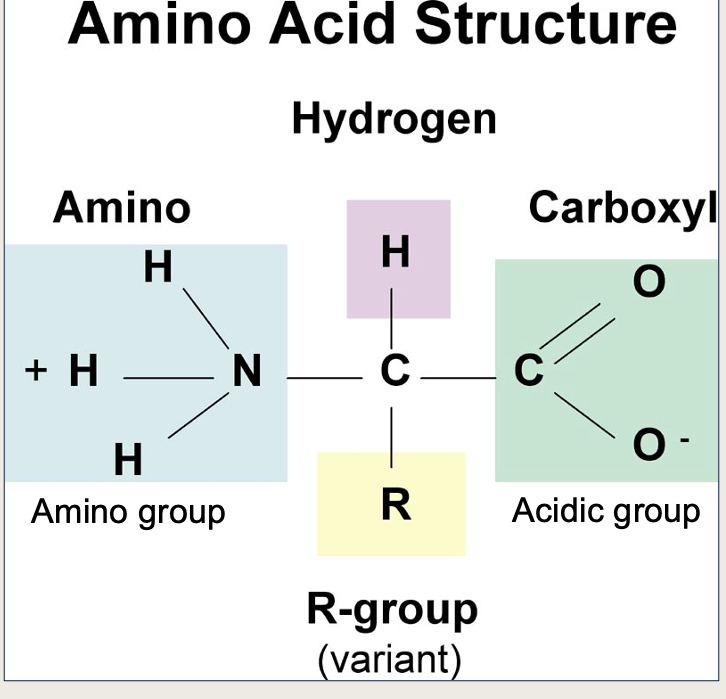
Define polar and non-polar
Polar = an unequal sharing of electrons between atoms
non-polar = an equal sharing of electrons between atoms
Where do essential amino acids come from?
come from food
How is a peptide bond made?
Amino acids are held together by peptide bonds generated by ribosomal peptidyl transferase
Peptide bonds are formed by a condensation reaction to form a covalent bond
The amino acids are joined in a genetically determined sequence
Is the amino group positive or negative?
Positive
Is the carboxyl group positive or negative?
Negative
True or false - A single polypeptide can only have one secondary structure
false
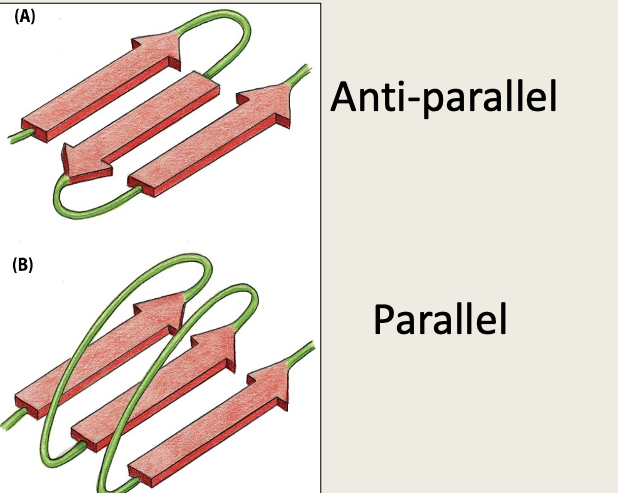
B pleated sheet - parallel and anti-parallel
Describe Alpha-helix
Polypeptide backbone folded into a spiral (5-20aa in length)
Shape forms due to hydrogen bonds forming between carbonyl groups and an amino group 4aa along the chain
3.6 amino acids per turn
Side chains face outwards
Describe interactions between the phospholipid bilayer and alpha helix
Hydrophobic side chains interact with the hydrophobic hydrocarbon tails of the phospholipids
The hydrophilic backbone is shielded from the lipid environment of the membrane
Hydrophilic parts of the backbone form hydrogen bonds between carbonyl groups and amino groups
Describe beta pleated sheet
flat structure (sheet)
laterally packed β strands
each β strand 5-8 amino acids
Shape forms due to hydrogen bonds forming between carbonyl groups and an amino group of a neighbouring chain
side chains extend above and below β-sheet
3 other secondary structures
Turns
Loops
Random coils
Describe turns
are 3-5 amino acids long forming a sharp bend redirecting polypeptide backbone
Describe loops
have hydrophilic residues and are found on the surface of proteins
Describe random coils
are polypeptide chains with random configuration
3 types of tertiary structure
Myoglobin
TNF alpha
Barrels
Describe barrels
first and last strand form hydrogen bonds forming a barrel
Describe beta barrells
Beta barrels often found in bacterial membrane proteins and some proteins in human mitochondria; they often allow ions to diffuse across membranes.
2 or 3 α-helices wind around each other forming a coiled coil.
Hydrophobic amino acids line up together where the helices meet. This provides structural strength to the protein.
Examples are keratin and collagen
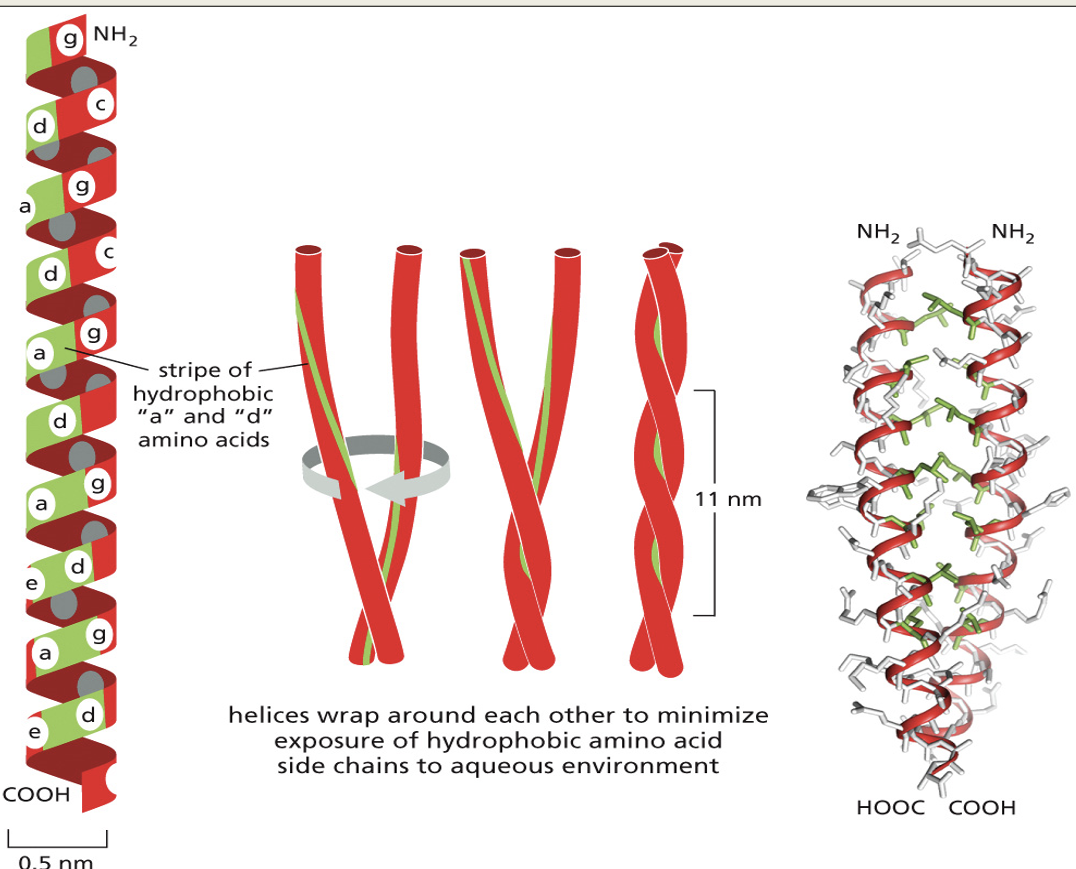
Side chain interactions
The peptide backbone is very flexible and allows rotation and folding.
This is stabilised by weak non-covalent bonds (hydrogen bonds when a H sandwiched between 2 electron attracting atoms), electrostatic reactions (between charged groups) and van der Waals forces (when molecules (atoms) are very close to each other (due to fluctuating electrical charge)).
They are all weak bonds but when many occur simultaneously that can hold the structure together. Stability of the protein is determined by how many of these bonds form.
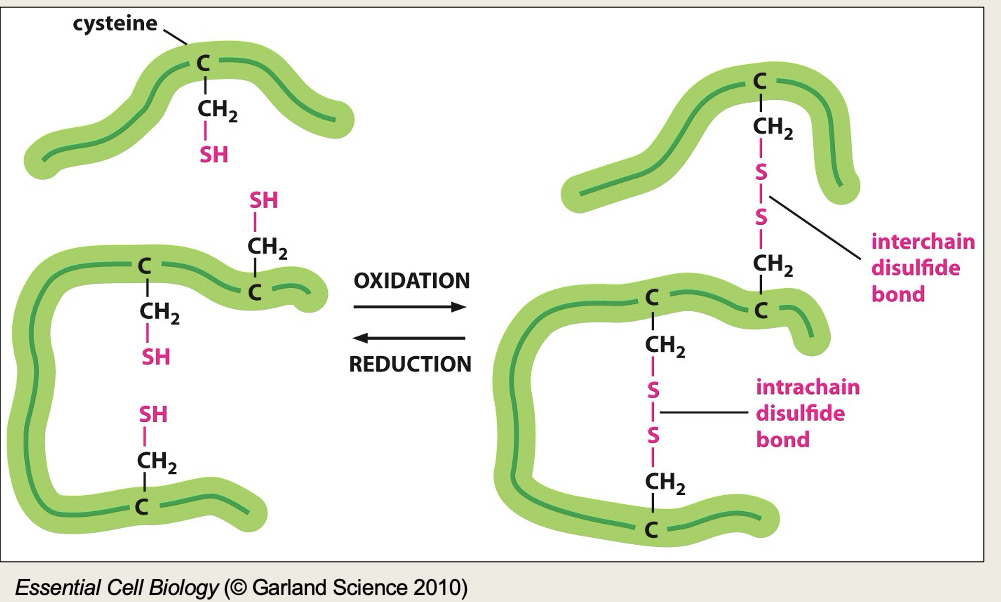
Describe disulphide bonds
Tertiary structure can be stabilised with covalent cross linkages. The most common is a disulfide bond between cysteines that are next to each other in the folded structure. They are formed by an enzyme in the endoplasmic reticulum.
They give strength but do not alter the shape.
Typically present in excreted proteins
Di-sulphide bonds are covalent bonds often seen in extracellular proteins e.g in saliva, they don’t often occur in cytosolic proteins due to reducing agents in the cytosol.
Needed where proteins may face pH changes.
Performed by enzymes in the endoplasmic reticulum - protein disulphide isomerase family (PDI).
Describe protein domains
Small proteins may fold into a single globular molecule.
Larger polypeptides may fold into two or more domains, each with their own 3D shape, separated by short linker chains.
Each domain of a protein may play a distinctive role in its function e.g. spanning the plasma membrane (receptor) or DNA-binding (transcription factors)
Domains often have a distinctive sequence motifs that enable their function to be predicted.
How is the quaternary structure formed?
hydrophobic interaction
What does haemoglobi consist of?
Haemoglobin is formed of 2 alpha subunits and 2 beta subunits
Binding of oxygen to one of the non-protein haem groups causes a conformational change that allow oxygen to bind more readily to the other heme groups
negatively charge carboxyl
Glutamic acid
Aspartic acid
Positively charged n group
Lysine
Arginine
histidine
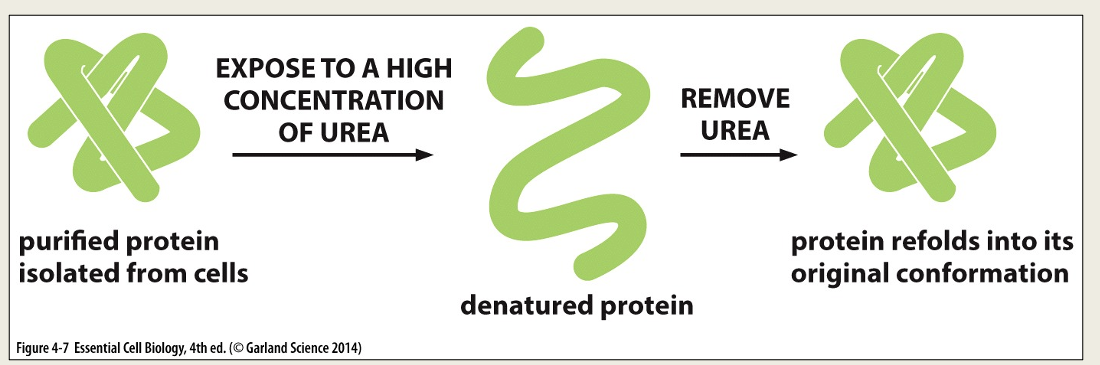
Protein denaturation
Proteins can be unfolded (denatured) by adding solvents (e.g. acids, bases) that disrupt the non-covalent interactions.
The protein loses its secondary and tertiary structures.
If the solvent is removed the protein will refold (renature) to the same shape as it started with.
Heat disrupts non-covalent interactions denaturing the albumin proteins which then aggregate and form new disulphide bonds altering the structure.
What is a folding funnel?
Proteins spontaneously fold into a three-dimensional conformation of the lowest free energy.
The folding process is energetically favorable. It releases heat and increases the disorder in the universe
Define energy and entropy
Entropy is a measure of disorder or randomness (number of possible ways to arrange the protein)
Energy is the potential energy available (due to movement, rotations and formation of bonds between atoms)
Describe molecular chaperones
Molecular chaperones – proteins that bind to the partially folded polypeptide chains and assist them in folding
Two classes include heat shock proteins and chaperonins
Chaperones: do not change the final 3D structure (native structure) but they do speed up the folding process, prevent protein aggregation and reduce non-productive intermediates
Called heat shock proteins as their expression is increased at elevated temperatures
Molecular chaperones are needed to help keep some proteins from misfolding on their way down the folding funnel, preventing them from straying into deep free energy wells (misfolded states).
Chaperones will bind the unfolded protein as it is made, shielding hydrophobic amino acids during the folding process.
Describe Post translational modifications (PTM)
A processing event resulting from proteolytic cleavage or the covalent addition of a modifying group.
Over 200 PTMs have been characterised.
PTMs modulate the function of most proteins by altering their activity state, localization, turnover, and interactions with other proteins.
Describe phosphorylation
Addition of a phosphate to specific amino acids regulating the activity of the protein
Describe glycosylation
Addition of carbohydrates to specific sites on the protein
N-linked glycans attached to a nitrogen of asparagine or arginine side-chains. N-linked glycosylation requires participation of a special lipid called dolichol phosphate.
O-linked glycans attached to the hydroxyl oxygen of serine, threonine, tyrosine, hydroxylysine, or hydroxyproline side-chains, or to oxygens on lipids such as ceramide
phosphoglycans linked through the phosphate of a phosphoserine.
Describe ubiquitination
Addition of ubiquitin can target the protein for destruction by the proteasome
Describe cystic fibrosis
Inherited disease of the secretory glands
DNA mutations that alter the conformation of a Cl- channel
Pulmonary obstruction due to thickened mucus in the small airways causing recurrent bacterial infections
Over 800 mutations detected
70% of patients share mutation at aa508
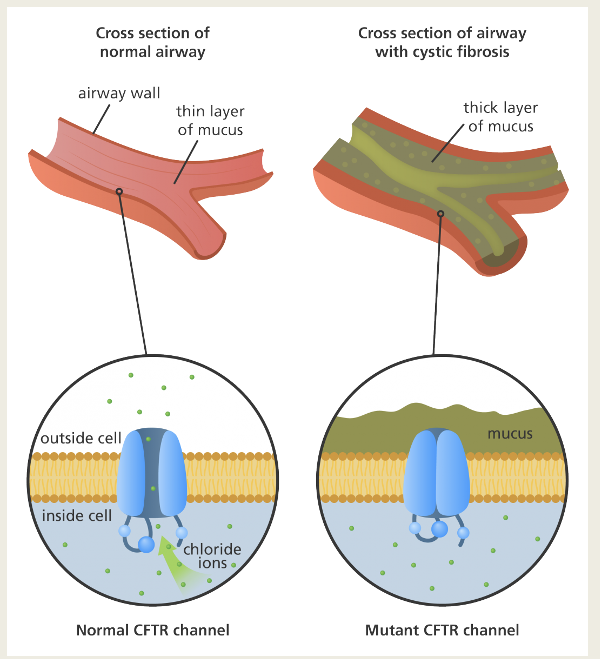
CFTR
All symptoms due to a single DNA mutation which causes the deletion of the amino acid Phenylalanine at position 508
Mutant CTFR protein becomes stuck in the endoplasmic reticulum leading to reduced chloride conductance out of the cells
ATC and ATT both code for isoleucine but the phenylalanine becomes deleted.
Lumacaftor is a small molecule drugs that can bind to and stabilise the protein of the Phe508 deletion gene mutation, meaning more CFTR can be trafficked to the cell surface.
AD accounts for 60% to 70% of dementia diagnoses
Most cases are sporadic, caused by a combination of genetic, lifestyle and environmental factors, with <1% of cases classed as familial AD, which is inherited
The disease is characterised by progressive memory loss and cognitive decline
Misfolding of amyloid beta causing amyloid beta plaques and tau aggregates causing intracellular neurofibrillary tangles (NFTs)
Misfolded proteins cause neuronal cell death
The build-up of amyloid beta plaques and NFTs cause irreversible neuronal cell death
Initial damage is in the areas of the brain associated with memory formation
Over time the damage becomes more widespread leading to shrinkage of the brain
Neuronal cell death is thought to be via necroptosis, a type of regulated cell death similar to necrosis.