Genetics chapter 7 - Bacteria and Viruses
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
50 Terms
Reasons for bacteria being a good organism
Rapid reproduction
Large progeny
Haploid genome allows all mutations to be expressed
Asexual reproduction
Small genome
techniques available to isolate and manipulate genes
medical importance
Type of bacteria
Prototrophic: wild type bacteria that can synthesize inorganic materials and survive on its own
Auxotrophic: mutant type bacteria that lacks enzyme responsible for synthesizing an essential molecule, requiring a complete medium
Techniques for studying bacteria
Minimum medium: only contains essential nutrients for prototrophic bacteria
Complete medium: Contains nutrients essential for all bacteria including auxotrophic
Difference between single colony and multiple colonies (in terms of genes)
Single colony: All progeny in a colony are genetically identical
Multiple colonies: One colonies genome may be different than another’s
Lab test for mutation steps
grow bacteria in test tube by placing into inoculate medium
After division, pipette onto growth medium
Spread and after 1-2 days colonies form
use velvet to stick and transfer colonies onto two mediums, one lacking amino acid, and other with it
Compare results (see if any colonies are missing to identify a mutation)
Bacterial genome
Bacterial DNA: mostly singular and circular chromosome
Plasmid: Extra, small circular DNA. Contain epistomes (f factors)
Plasmid conflict
A single plasmid can carry many genes. They can carry multiple different types of antibiotic resistant genes.
Use of antibiotics promoting natural selection
Bacteria containing a plasmid that has a gene for resistance against an antibiotic will survive when antibiotics are used. These bacteria will produce and more bacteria with the plasmid containing antibiotic resistant gene will spread.
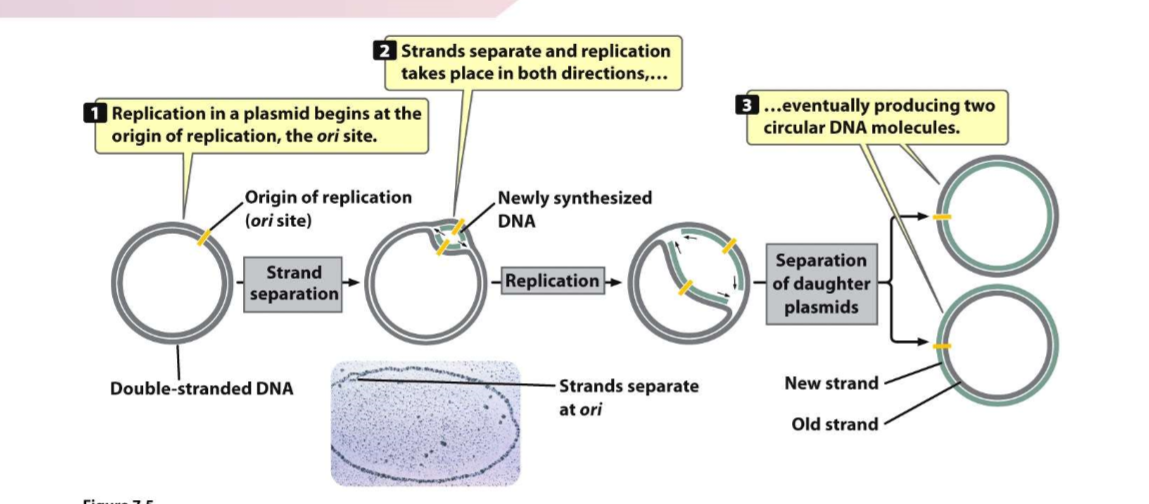
Replication process for plasmid
Replication begins at ori site
strands separate and replication begins in both directions for each strand
Two daughter cells will be produced, possibility of new strand inside or outside
F factor
Fertility factor, DNA segment that allows for exchange of genes through conjugation
3 types of gene function for f factor
Genes for regulating plasmid replication
genes that regulate plasmid transfer to other cells
Genes for regulating insertion of plasmid into bacterial chromosome
Experiment to identify if bacteria exchange genes and result
separate bacteria that are auxotrophic for opposite strains
Place both in a minimum medium where they cannot grow individually
Formation of colonies indicates that bacteria exchanged info to make completely prototrophic bacteria
Result: Bacteria do exchange genetic info
Experiment to see if direct contact is needed and result
Two bacteria, auxotrophic for differing segments, are placed in the same medium
A filter is present to allow medium to cross but not the bacteria
After, the bacteria are pipetted onto minimal medium and none survive
Result: Bacteria need direct contact to exchange genetic info
Three types of bacteria genetic exchange
Conjugation: direct contact between bacteria in order to exchange genetic info
transformation: Cell lysis and a new bacteria picks up free gene fragment
Transduction: Virus lysis cell and takes gene fragment, then virus infects another bacterial cell and the gene fragment is passed on
Different F factors
F+: plasmid is separate DNA molecule
F-: plasmid is absent
Hfr: Present and integrated into bacterial chromosome
F’: Plasmid is separate DNA molecule containing some bacterial genes
F factors roles in conjugation
F+: donate plasmid
F-: receive plasmid
Hfr: high frequency plasmid donor
F’: donate plasmid
Conjugation
Direct transfer of one plasmid strand from F+ cell, through conjugation pilus (sex pilus), into receiving F- cell. (Not reciprocal)
Formation of Hfr cell
crossing over between F factor on plasmid and bacterial chromosome results in integration into bacterial chromosome
Merozygote
Partial diploid bacterial cell containing F plasmid carrying some bacterial genes
How does merozygote form
F’ transfers f plasmid containing some chromosomal gene to F- and then variations of same gene can arise
Result of conjugation between cells with different f type
F+ x F- : Two F+ cells (F- becomes F+)
Hfr x F- : One Hfr and one F- (F- might have genetic change as crossing over can occur from loose Hfr fragment and bacterial chromosome
F’ x F- : Two F’ cells (F- becomes F’ and partial diploid/merozygote)
gene mapping with conjugation
Distance between genes is measure using time it takes to transfer DNA from Hfr to F-
Interrupted mating
Experimental way to measure gene location using conjugation
Transformation
A bacterium takes a DNA fragment from medium. Recombination then takes place to integrate into the bacterial chromosome
Component cell
Cells that prepares to take up DNA from medium during transformation when under stress
Types of transformation cells
transformant: Cell that receives genes and integrates
cotransformed: Cells transformed by two or more genes
transformation steps
Cell takes one strand of DNA from medium and other strand is hydrolyzed
the strand attaches to bacterial chromosome and recombination occurs
When the cell replicates, original strand will produce a daughter cell with original bacterial genome and other will contain two transformed strands
Laboratory use of transformation
heat shocking cell produces component cell which is more readily able to take up foreign DNA
Transformation efficiency
Probability that cell will take extracellular DNA and express genes encoded in it
factors affecting transformation efficiency
Plasmid size
DNA type
cell genotype
transformation method
cell growth rate
gene mapping using co transformation
Transformants that take up two or more genes from cell fragments will exhibit the trend:
Closer two genes are = higher rate of co-transformation
Bacterial defense mechanisms
reduce expression of receptors viruses need to attach to
Secretion of polysaccharides to limit infection
Block viral replication
CRISPR-Cas systems (main focus)
CRISPR-Cas system
Recognizes and remembers DNA of specific pathogens. Acts as bacteria’s immune system
CRISPR - Clustered regularly interspaced short palindromic repeats.
(repeated sequences separated by spacers that are the same forward or backward)
CRISPR-Cas explained
Viral DNA is copied by CRISPR and copied viral sequences are made so that when viral DNA comes back, Cas can recognize it and cut it out.
How are double stranded DNA breaks repaired?
NHEJ (non homologous end joining) - Brings two ends of a break together
HDR (Homologous directed repair) - Adds donor DNA to break
Problem with NHEJ
Often results in nucleotide insertions or deletions
Virus
replicating structure of DNA/RNA and a protein coat
Virulent phage
will always kill host cell and reproduce through lytic cycle
temperate phage
inactive prophage whose DNA integrates into bacterial chromosome
Lytic cycle (6 steps)
1) attachment - virus locks onto bacterial cell
2) injection - virus injects DNA into bacterial cell
3) Biosynthesis - bacterial cell stops its own work and instead follows genome of virus
4) Assembly - Viral parts begin to assemble themselves into many new viruses inside cell
5) Lysis - Cell breaks open
6) Release - Viruses are released after cell lysis
Lysogenic cycle
Instead of degrading host cell chromosome, viral DNA is incorporated into it. Temperate prophages enter this state
Transduction
Use of phages to map bacterial genes
Bacterial viruses carry bacteria DNA from one bacteria to what is often the same or similar bacterium
Transduction steps
Virus injects DNA into bacteria
Phage DNA and bacteria DNA are fragmented
Phages with viral DNA and bacteria DNA are produced
Bacteria cell lyses, releasing viruses
Virus with bacteria DNA injects DNA into different bacterial cell
DNA gets implemented into bacteria chromosome
Transduction mapping
Same trend as transformation:
Closer two genes are = Higher rate of cotransduction
(two genes close together are more likely to be on same DNA fragment that generated Virus conatins)
Equation for determining position of a gene on a phage chromosome
RF = recombinant plaques / total plaques
RNA viruses
Retroviruses: RNA virus integrated into hosts genome (not integrated into chromosome directly)
Reverse transcriptase: synthesizes DNA strand from RNA template
Provirus: RNA genome incorporated into hosts chromosomes
Process of Retrovirus
Release reverse transcriptase: synthesizes DNA strand from RNA template
The RNA genome can then be incorporated into hosts chromosomes called a Provirus
Important virus genes
gag - encodes proteins that make viral proteins
Pol - encodes reverse transcriptase and enzyme integrase
Env - encodes glycoproteins found on viral surface
Two protein types for influenza
HAS - Hemaggutinin
Amindase (NA)
Antigenic shift vs Antigenic drift
Antigenic shift - Major genetic change in influenza that results in a new sub-type of the virus
Antigenic drift - Small genetic mutations in viruses genes