biochem chapter 3 - working with proteins
1/122
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
123 Terms
a protein is defined by its sequence of _____ _____
each cell type has a distinct set of _____
the entire complement of proteins is called the ______
amino acids
proteins
proteome
~____ protein-coding genes (HGP)
~____ have been detected in at least one tissue
~____ are present in all tissues
~____ are essential
~20,000 protein-coding genes (HGP)
~18,000 have been detected in at least one tissue
~10,000 are present in all tissues
~2,000 are essential
~20,000 proteins are ____
~18,000 have ____
~10,000 proteins are ____
~2,000 are ____
~20,000 protein-coding genes (HGP) TOTAL #
~18,000 have been detected in at least one tissue
~10,000 are present in all tissues
~2,000 are essential
Proteome is actually much larger than genome because of:
____
____
____
post translational modifications
synthesis intermediates or precursors
alternative splicing
protein functionality is enhanced through _____ _____
ligand binding
lipoproteins
nonprotein part: ____
examples: ____
lipids
blood lipoprotein complexes, HDL, LDL
nucleoproteins
nonprotein part: ____
examples: ____
RNA/DNA
Ribosomes, chromosomes
glycoproteins
nonprotein part: ____
examples: ____
carbohydrate groups
immunoglobulins, LDL receptor
metalloproteins and metal-activated proteins
nonprotein part: ____
examples: ____
Ca+,K+,Fe2+,Zn2+,Co2+, others
Metabolic enzymes, kinases, phosphates, others
hemoproteins
nonprotein part: ____
examples: ____
heme group (carries O2)
hemoglobin, cytochromes
flavoproteins
nonprotein part: ____
examples: ____
FMN, FAD
electron transfer enzymes
peptides are short polymers of amino acids. individual amino acid units are called ___.
residues
2 residues - dipeptide
3 residues – tripeptide
12-20 residues – ____
>20 residues – ____
>50 residues – ____
oligopeptide
polypeptide
protein
protein sequences are unique and are conventionally read ____.
N → C (amino terminus→ carboxyl terminus)
preparative approaches result in protein to ____.
analytical approaches ____.
preparative approaches result in protein to “work with”.
analytical approaches reveal something about the protein.
what protein properties do both preparative and analytical proteins make use of?
binding
charge
hydrophobicity
size
solubility
density
the 3 steps for purifying proteins is
first step = break open tissue or microbial cells
– ____ = releases proteins in solution
second step = ____ = separate proteins into fractions based on size or charge
– “____” = lower solubility of proteins in salt to selectively precipitate proteins
third step = ____ = use semipermeable membrane to separate proteins from small solutes
crude extract
fractionation, salting out
dialysis
when purifying, what is the goal?
highest purity with the least amount of steps
more steps leads to less yield and more purity
centrifugation is what kind of process?
differential(non-equilibrium) - because its separated due to force
during centrifugation, particles ____ from solution at different speeds.
______ particles sediment faster.
PELLET
larger and denser
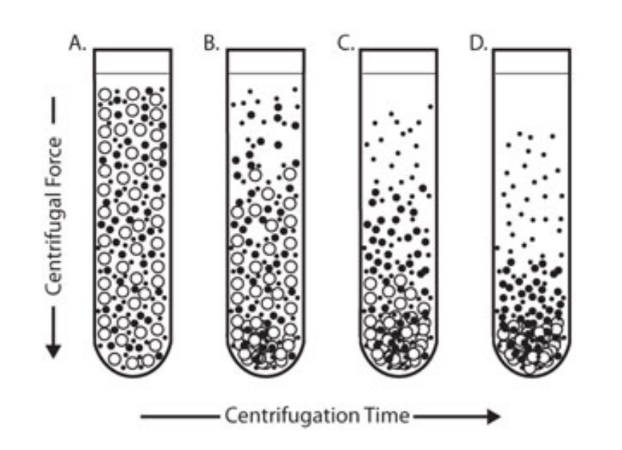
centrifugation is used for: (2)
harvesting cells(one spin)
separating organelles using a series of spins or cell fractionation
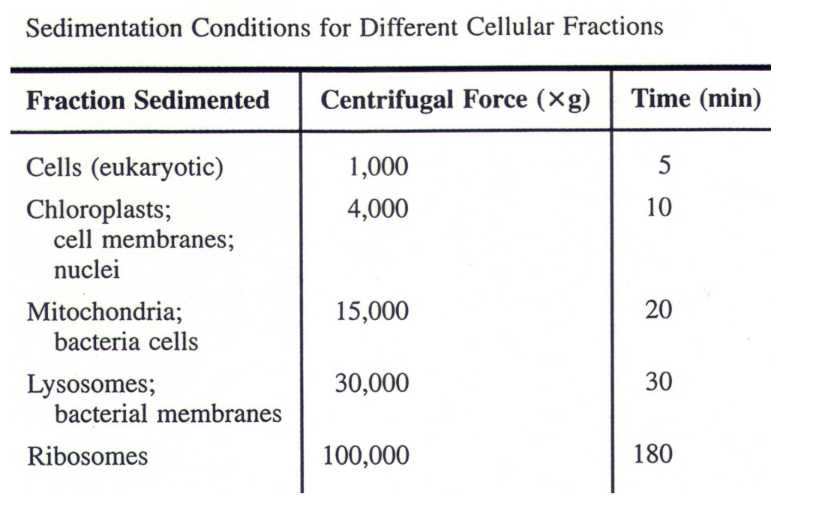
protein solubility is influenced by ___ and ___
pH, ionic strength
proteins are least soluble when?
when are they the most soluble?
least - at their pI (neutral)
most - away from pI, when charged
salting in vs salting out on solubility
salting in - low salt concentrations improve solubility
salting out - high concentration decrease solubility
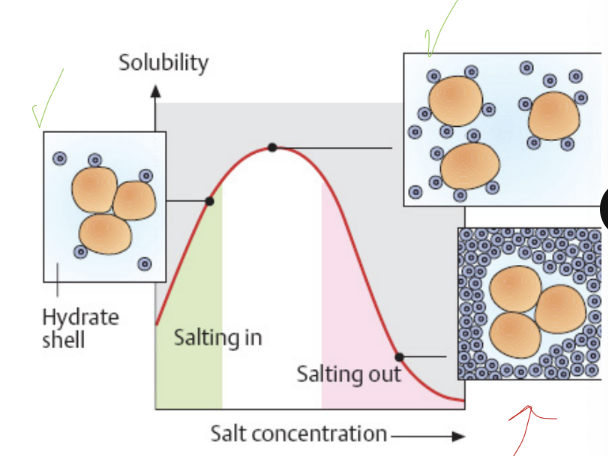
during ___ ____adding the right amount of salt(___ ___) can precipitate contaminants or molecules of interest.
salting out ; ammonium sulfate
S100 fraction and S100 proteins are _____.
solubility in 100% ammonium sulfate at neutral pH
during ____ molecules separate based on their interactions with the stationary phase
liquid chromatography
during liquid chromatography a column has a ____ phase(liquid) and a ____ phase (resin).
molar absorbance at ____
during liquid chromatography a column has a mobile phase and a stationary phase (resin).
molar absorbance at 280 nm
stationary phase of liquid chromatography
are made up of ____
they are mechanically ____ (high pressures)
chemically ____
____
____ support for chemical groups
are made up of hydrated polymers (polysaccharides)
they are mechanically stable (high pressures)
chemically inert (don’t react with sample components)
cheap
physical support for chemical groups
what are different stationary phases made of? (3 examples)
agarose
amylose
cellulose
column chromatography has 2 steps:
First step: The buffered solution (mobile phase) migrates through the porous solid material (stationary phase) to equilibrate the column.
Second step: The buffered solution containing the protein mixture is added and migrates through the solid phase, allowing separation based on each protein’s interactions with the stationary phase.
protein properties affect _____ of column chromatography
migration rate
____ separates based on sign and magnitude of the net electric charge
ion-exchange chromatography (IEX)
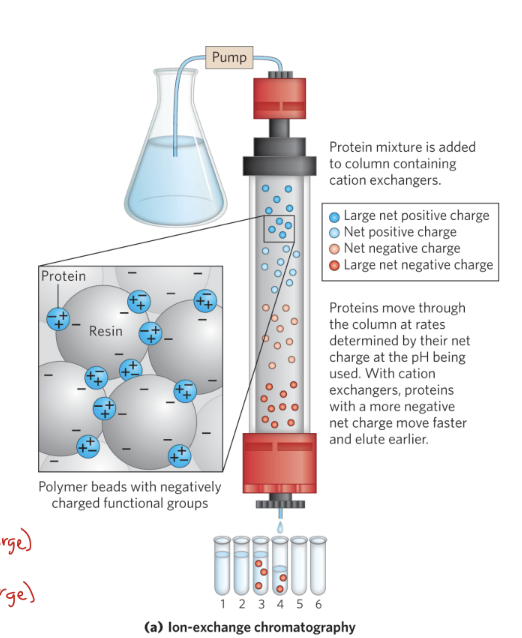
during IEX, pH and concentration of free salt ions affect ____
protein affinity
IEX involves
cation exchangers → ____ charged resin binds ____ charged proteins.
anion exchangers → ____ charged resin binds ____ charged proteins.
cation exchangers → negatively charged resin binds positively charged proteins.
anion exchangers → positively charged resin binds negatively charged proteins.
during IEX, proteins are loaded in ___, which means there is low salt, and use gradient of ___ salt to elute(release from column)
salt ex: NaCl, KCl
low
increasing
what does a chromatogram represent in IEX and how is it interpreted?
A chromatogram is a graph showing absorbance at 280 nm (y-axis) versus elution volume or time (x-axis).
Peaks correspond to proteins or other molecules eluting from the column.
Early peaks = weakly bound or unbound proteins (elute at low salt).
Later peaks = strongly bound proteins (require higher salt to elute).
The salt gradient (NaCl or KCl) is often shown on a secondary axis, rising as elution progresses.
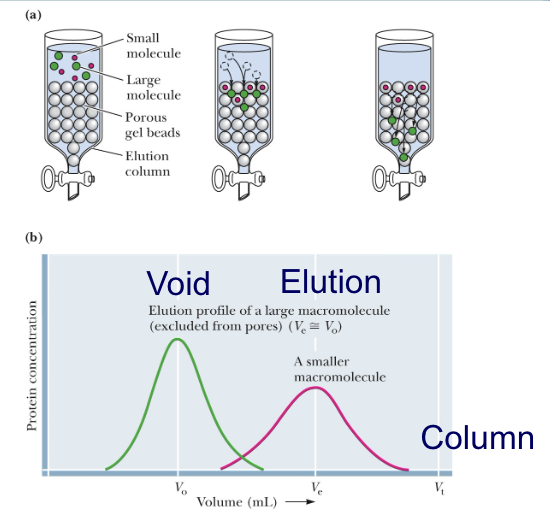
what are other names for size-exclusion chromatography(SEC)?
also called gel filtration chromatography
what’s the order of protein emergence during SEC?
large proteins emerge from the column before small proteins
small proteins have to go through the beads but the big ones just pass freely around beads
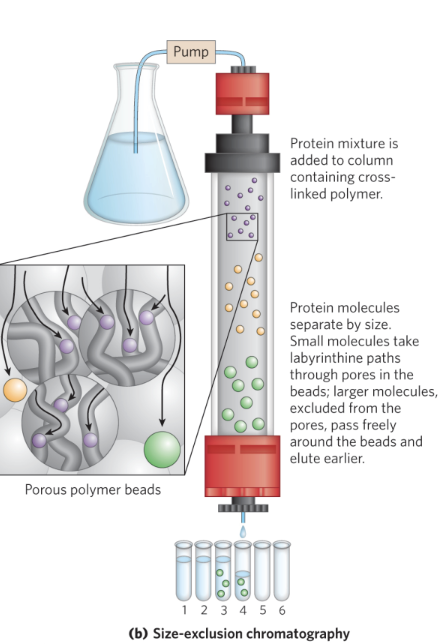
which molecules are voided during SEC?
the larger macromolecule is is voided because it never actually travelled through the resin
affinity chromatography separates based on ___ affinity
binding
affinity chromatography properties
resin carries ligands that bind the target protein
high specificity! >95% purity in a single step
to isolate the target protein: elute with a solution that disrupts the protein-ligand interaction, typically an excess of competing ligand
examples of affinity chromatography:
GST tag binds GSH
His tag binds Ni-NTA
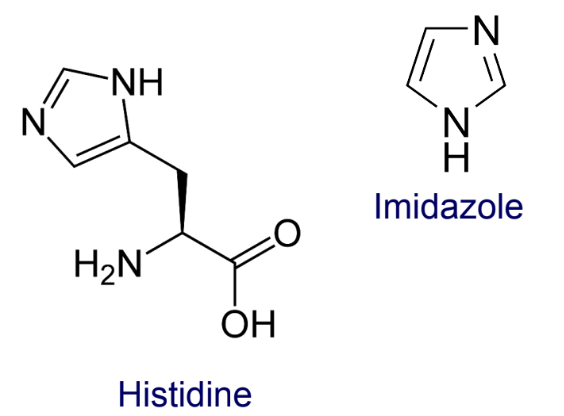
how does Ni²⁺ affinity chromatography purify His-tagged proteins?
The resin contains nitrilotriacetic acid (NTA) groups that coordinate(bonds) Ni²⁺ ions.
The target protein is engineered with 6–8 histidine residues (His-tag).
The imidazole rings of histidines bind to the Ni²⁺ on the resin.
After washing away unbound proteins, the His-tagged protein is eluted by adding imidazole, which competes with histidines for Ni²⁺ binding sites.
how does hydrophobic interaction chromatography (HIC) work?
Hydrophobic groups covalently-linked to polar resin (HIC)
Reverse of salting out
Load in high [salt] and use gradient of decreasing [salt] to elute
hydrophobic proteins don’t like water, so high salt → high solubility
where in the cell are most hydrophobic proteins?
the inner cell membrane, so lots of transmembrane proteins are purified by HIC
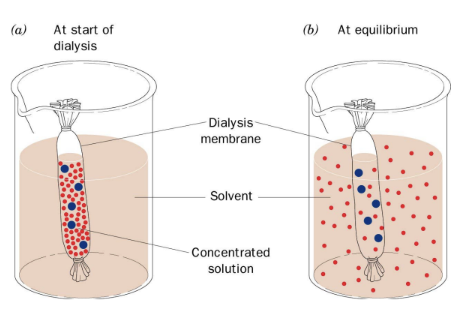
what are the principles of dialysis
diffusible solutes in the dialysis bag equilibrate across the membrane
membrane pores should be smaller than the size of the protein
what are the principles of ultrafiltration
Upper and lower chambers separated by a semipermeable membrane
Centrifugation or vacuum is used to force water and small solutes through the membrane leaving concentrated target protein in the upper chamber
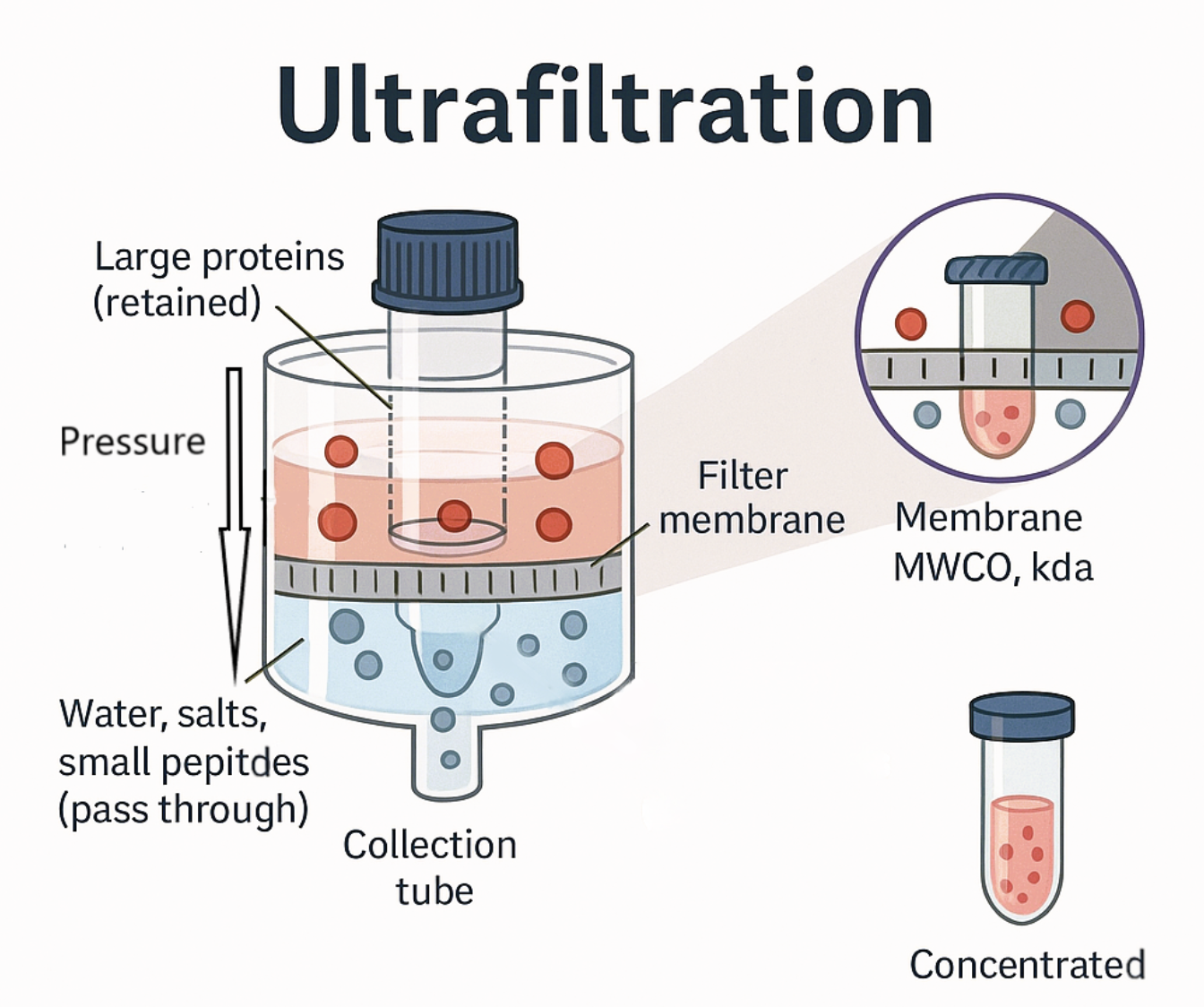
is dialysis or ultrafiltration faster?
ultrafiltration is faster than dialysis
because dialysis is passive diffusion while ultrafiltration is force driven by pressure of a vacuum actively pushing water and small solutes
sequential purification steps increase/decrease sample size
decrease
how to calculate purification factor?
purification factor = (final specific activity) / (starting specific activity)
final specific activity: after a purification step
starting specific activity: in the crude extract (or previous step if cumulative)
meaning: How many times purer the protein is compared to the crude sample
how to calculate percent yield
%final activity / %starting activity activity = percent yield
___ visualizes and characterizes purified proteins
electrophoresis
what can electrophoresis be used to estimate?
number of different proteins in a mixture
degree of purity
isoelectric point (pI)
approximate molecular weight
what is beer lambert’s law and what is it used to determine?
used to determine protein concentration
A = εcl
A=absorbance at 280nm
ε=extinction coefficient at 280nm (will be given)
c=concentration
l=path length

What are the 2 types of Electrophoresis?
Native PAGE
SDS-PAGE
What are the characteristics of Native PAGE?
Proteins remain folded and functional
Migration determined by size, charge, and shape
Cannot reliably estimate molecular weight, because shape and charge effect mobility, not just weight
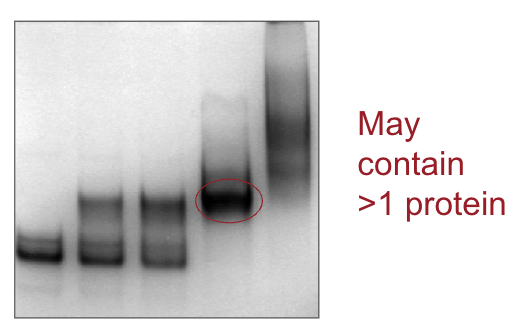
What are the characteristics of PAGE?
General method to separate proteins using an electric field through a polyacrylamide gel.
Proteins migrate from cathode (-) to anode (+).
‘See’ proteins by using a stain
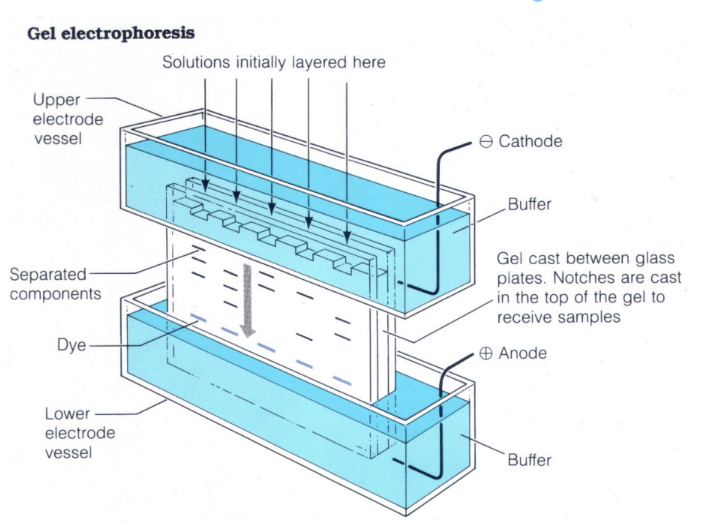
how does electrophoresis separate and visualize proteins?
Uses cross-linked polyacrylamide gels (PAGE).
Proteins migrate based on their charge-to-mass ratio under an electric field.
Migration is from cathode (-) to anode (+).
Proteins are visualized using Coomassie blue dye, which binds to proteins.
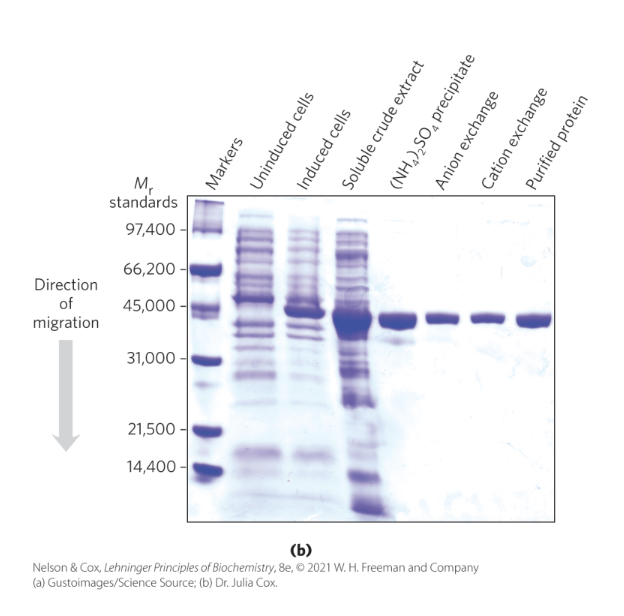
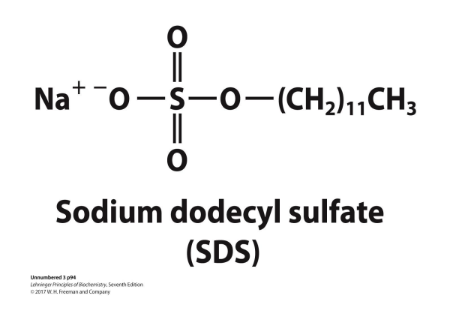
what is SDS and why is it important in SDS-PAGE?
sodium dodecyl sulfate (SDS) = a detergent
binds and partially unfolds proteins
gives all proteins a similar charge-to-mass ratio
electrophoresis in the presence of SDS separates proteins by molecular weight
because same MW but diff charge travel differently, once all charges are same(negative), move only based off of MW
smaller proteins migrate more rapidly
how to calculate electrophoretic mobility(μ)?
μ = V/E = Z/f
μ = electrophoretic mobility
V = velocity
E = electrical potential
Z = net charge
f = frictional coefficient
how to estimate the molecular weight of a protein?
plot of log Mr of marker proteins vs relative migration during electrophoresis = linear
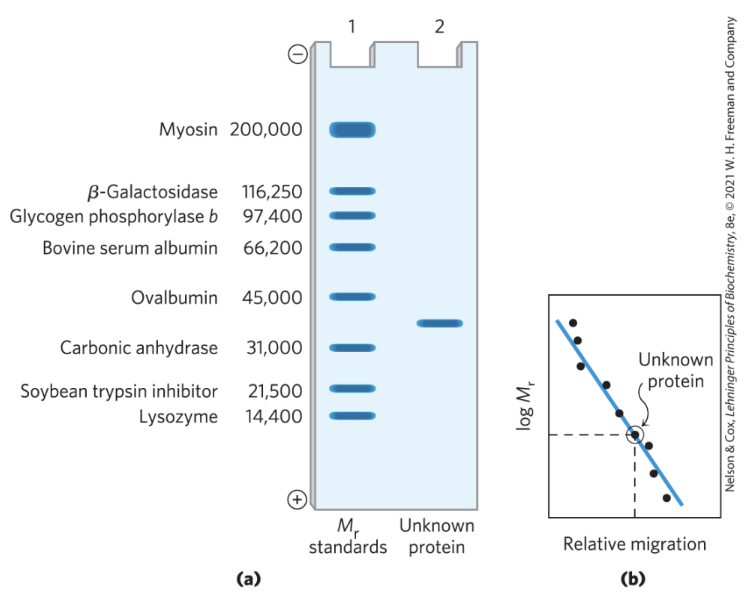
what happens if an oligomeric proteins is separated on a SDS page (e.g. homotetramer of 80kDa)?
it will dissociate and run as a 20kDa monomer
using ____ to determine the pI of a protein
isoelectric focusing
**not used anymore
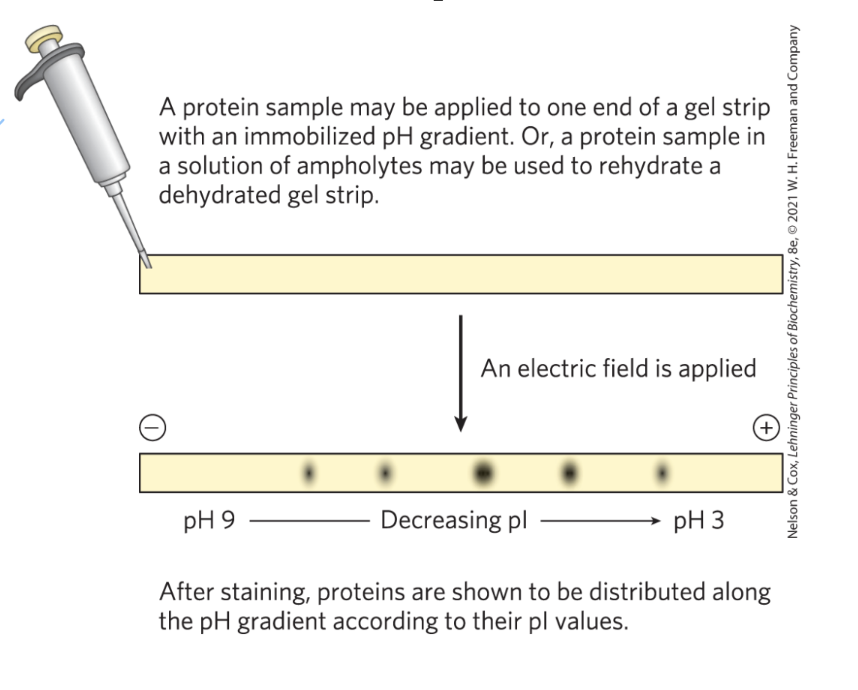
two-dimensional electrophoresis characteristics
permits resolution of complex protein mixtures of proteins
more sensitive than individual methods
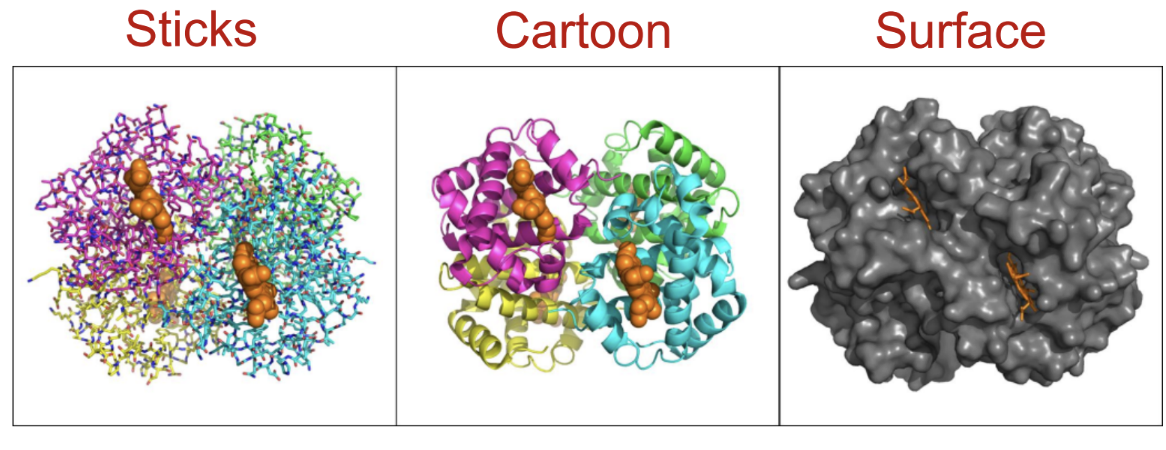
hemoglobin is a ____ - it has two alpha chains and two beta chains
heterotetramer
what is protein databank and pymol?
Protein Data Bank (PDB):
A public database of experimentally determined 3D structures of proteins, nucleic acids, and complexes.
PyMOL:
A molecular visualization software used to view and manipulate 3D protein structures..
what are the 2 general shape classifications for proteins
fibrous
globular
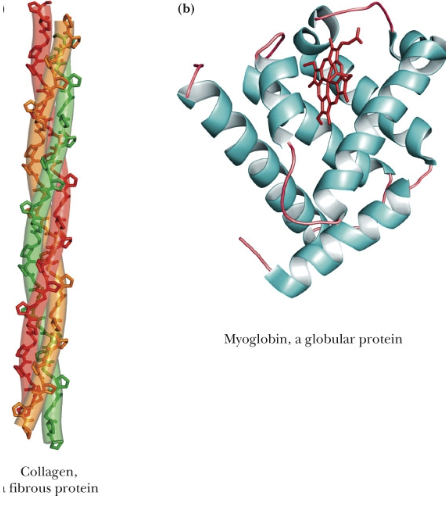
fibrous vs globular protein shapes
fibrous proteins have an extended structure and are structural proteins like collagen, elastin, and keratin
globular proteins are sphere-like in structure. most cytosolic enzymes have this shape.
because cytosol is packed and globular proteins take up less space than fibrous proteins
protein classification by solubility
soluble proteins:
hydrophilic residues on the surface, hydrophobic residues buried inside
easily dissolved in aqueous buffers
insoluble proteins (e.g., membrane proteins):
hydrophobic residues exposed on the surface
soluble in detergents
what are the four levels of structure in proteins?
primary structure - amino acids linked by covalent bonds in a polypeptide chain
secondary structure - recurring structural patterns like alpha or beta helices
tertiary structure - 3D folding of polypeptide
quaternary structure - 2+ polypeptide subunits
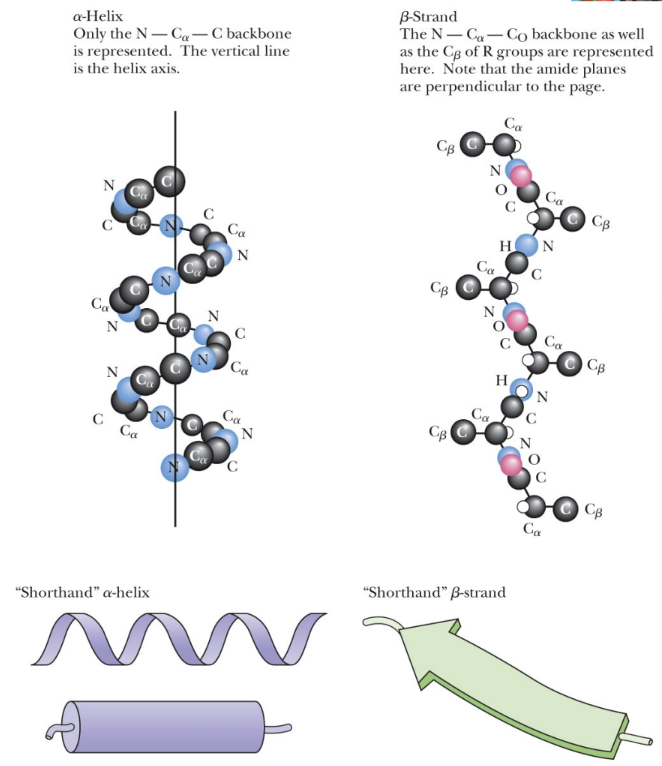
what are the two principal secondary structure found in proteins?
α-helix
β-pleated strand
sequencing of the first protein
fred sanger was the first to sequence a protein
insulin - 1953
he went on to figure out how to sequence DNA
showed that all molecules of a protein have a fixed amino acid composition
what method is used to sequence proteins today?
mass spectrometry
what are the steps of protein sequencing?
separate non-covalently linked chains
break and block disulfide bonds
cleave proteins (fragmentation) into peptides using sequence specific proteases or chemicals
isolate and separate peptides
sequence peptides from N-terminus and C-terminus
repeat 3-5 with different cleavage until good overlap between fragments
assemble full sequence
edman degradation and carboxypeptidases in protein sequencing?
Edman Degradation: sequencing from N-terminal → C-terminal
Carboxypeptidases: sequencing from C-terminal → N-terminal
how do we separate non-covalently linked chains? (1st step of protein sequencing)
Overcome weak forces using:
high salt (ammonium sulfate)
chaotropes like 8 M Urea or 4-6 M Guanidine-HCl
interfere with H-bonds and reduce “the hydrophobic effect” a.k.a they disrupt water and allow otherwise buried hydrophobic residues to be soluble and thus exposed
how do we break and block disulfide bonds linked chains? (2nd step of protein sequencing)
to break:
use reducing agents and then modifiers
in the lab prevent with reducing agents such as TCEP, β-
mercaptoethanol and dithiothreitol (DTT)
to block from reforming:
because reaction between the newly reduced –SH groups to reestablish disulfide bonds is a likelihood, S-S reduction must be followed by –SH modification
modifiers prevent disulfides re-forming!!!
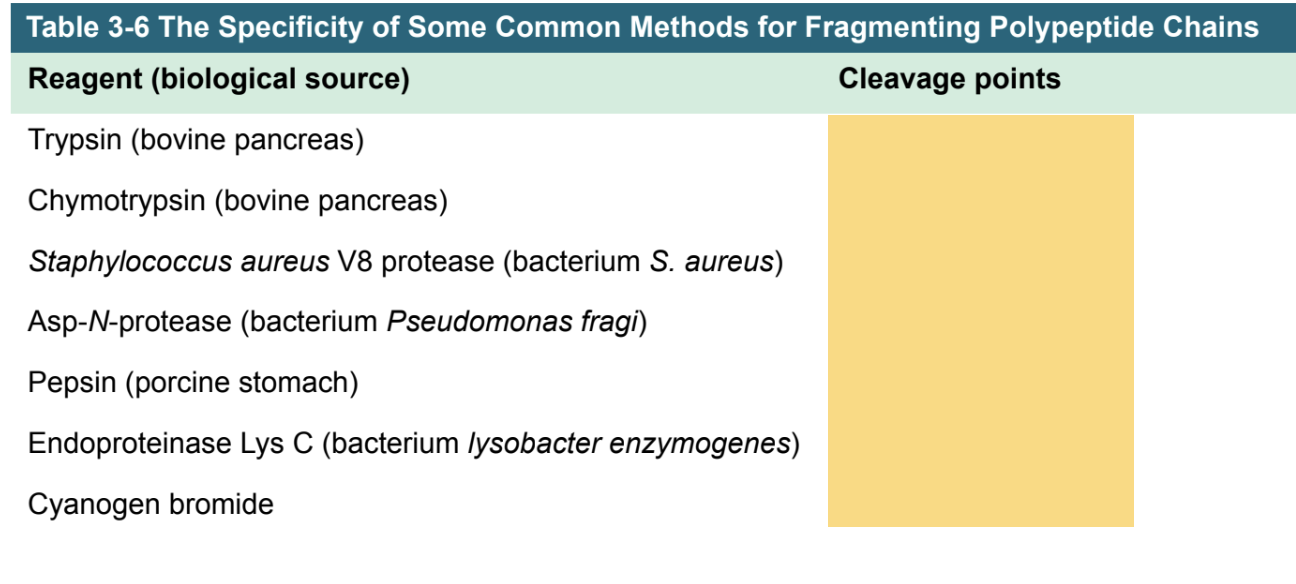
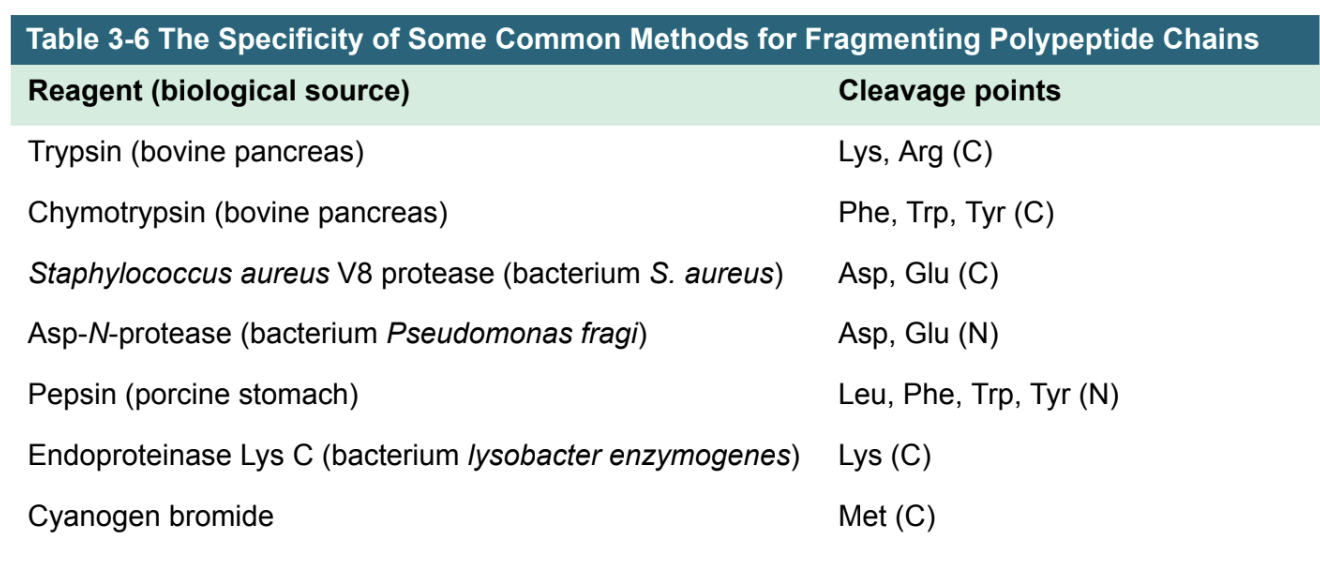
how do we cleave proteins into peptides w/proteases and chemicals? (3rd step of protein sequencing)
Protease | Cleavage Specificity | Notes |
|---|---|---|
Trypsin | After Arg (R) or Lys (K) | Cleaves on the C-terminal side of basic residues |
V8 Protease (Glu-C) | After Glu (E) in ammonium buffer; sometimes after Asp (D) in phosphate buffer | C-terminal cleavage |
Chymotrypsin | After Phe (F) or Tyr (Y) (aromatic residues) | C-terminal cleavage |
Pepsin | Preferentially cleaves aromatic and acidic residues in acidic pH | Works under low pH |
Thermolysin | Cleaves before Leu, Ile, Val, and other hydrophobic residues | N-terminal cleavage; prefers hydrophobic residues |
proteases = catalyze ____ of peptide bonds
proteases = catalyze hydrolytic cleavage of peptide bonds
___ immobilizes peptide on matrix via C-terminus, leaving the n-terminal free to react.
edman degradation
edman degradation
cycle 1 - identifies ____
cycle 2 - identifies ____
less efficient with ___
can do 50 cycles for a 100-200 aa protein
can do 10-20 cycles for a 2000 aa protein
cycle 1 - identifies N-terminal residues
cycle 2 - identifies N+1 residue
less efficient with large proteins
can do 50 cycles for a 100-200 aa protein
can do 10-20 cycles for a 2000 aa protein
reverse phase chromatography
resin contains hydrocarbon chains of ____ ____(C4, C8, C12)
longer chain is more ____
load in ____ solution (water) and elute with gradient of ____ solution (organic solvent such as acetonitrile)
separation based on ____
non-polar solutions often ____ proteins so more widely applied to ____
reverse phase chromatography
resin contains hydrocarbon chains of differing lengths (C4, C8, C12)
longer chain is more hydrophobic
load in polar solution (water) and elute with gradient of non-polar solution (organic solvent such as acetonitrile)
separation based on hydrophobicity
non-polar solutions often denature proteins so more widely applied to peptides
the seven step protein sequencing steps have been replaced with _____: (2 types of it)
• _____
• _____
mass spectrometry approaches:
• peptide mass fingerprinting (with databases)
• peptide sequencing using tandem MS
_____ = measure molecular mass with high accuracy
can sequence short amino acid sequences (20 to 30 amino acid residues)
can document the entire cellular proteome
mass spectrometry
short
proteome
mass spectrometry
separates molecules (in this case peptides) based on their ___
detects ___ or ___ molecules in the ___ phase
mostly ___
cannot detect uncharged molecules
___ or ___ ___ maintains covalent bonds
separates molecules (in this case peptides) based on their mass-to-charge ratio (m/z)
detects charged or ionized molecules in the gas phase
mostly protonation
cannot detect uncharged molecules
weak or soft ionization maintains covalent bonds
what are the 3 components of MS?
source: evaporate and ionize molecules in a vacuum to create gas phase ions e.g. ESI or MALDI
mass analyzer: Separate ions in space or time based on their m/z ratios
detector: measure the amount of ions with specific m/z ratios
source: ESI - ___
solution sample is sprayed as fine droplets from a glass capillary under a ___ ___ ___
often inline with ___ ___ ___
Electrospray Ionization
strong electric field
liquid chromatography (LC-MS)
what does an ESI spectrum look like?
x-axis: m/z units
y-axis: relative intensity
each peak are different charge states
tallest peak is the base peak
what is the base peak on an ESI spectrum?
The base peak is the tallest peak in a mass spectrum.
Represents the most abundant ion detected in the sample.
Its intensity is set to 100%, and all other peaks are measured relative to it.
Does not necessarily correspond to the molecular ion (the intact molecule); it could be a fragment or adduct that is most easily detected.
what is the second type of source for MS?
MALDI (matrix-assisted laser desorption ionization)
characteristics of MALDI
the matrix is a light absorbing substance that is excitable by a laser
sample is mixed with a matrix and dried on a surface
lasers ionize the matrix and facilitate the transfer of a proton
not inline with chromatography (off-line)
most molecules pick up only a single H+
limited mass range
what is the analyzer for MS?
TOF (time of flight) analyzer
TOF -
ion's mass-to-charge ratio is determined via a____. Ions are accelerated by an _____ of known strength.
acceleration results in an ion having the same ___ as any other ion that has the same charge.
the velocity depends on the _____ (heavier ions of the same charge reach lower speeds, although ions with higher charge will also increase in velocity).
ion's mass-to-charge ratio is determined via a time of flight measurement. Ions are accelerated by an electric field of known strength.
acceleration results in an ion having the same kinetic energy as any other ion that has the same charge.
the velocity depends on the mass-to-charge ratio (heavier ions of the same charge reach lower speeds, although ions with higher charge will also increase in velocity).
Peptide Mass Fingerprinting
identification of a protein based off the mass of its ____
can predict tryptic peptides of all proteins in a ____
___ and ___ databases
identification of a protein based off the mass of its tryptic peptides
can predict tryptic peptides of all proteins in a database
genomic and proteomic databases
peptide mass fingerprinting
1) Peptides are generated by ___ ___ using a specific protease
2) Peptide masses are determined using ___ or ___
3) ___ ___ ___ are calculated for each entry in the protein database using the same protease
4) A ___ is calculated to measure the fit between experimentally derived and calculated peptide masses
1) Peptides are generated by protein digestion using a specific protease
2) Peptide masses are determined using MALDI-MS or ESI-MS
3) Theoretical peptide masses are calculated for each entry in the protein database using the same protease
4) A ranking is calculated to measure the fit between experimentally derived and calculated peptide masses
T/F all peptides can be detected by MS
False
Why can’t all peptides be detected by MS?
Convoluted by peptides with similar masses but different sequences
Convoluted by post-translational modifications