6.3 Manipulating Genomes
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
33 Terms
What is the general name for the process that determines the order of nucleotides in a length of DNA?
DNA sequencing
Main technique for sequencing DNA
Sanger sequencing / chain termination technique
Outline how DNA is sequenced
Extract DNA
Cut DNA into fragments of various lengths
Amplify
Sequence DNA fragments
Place fragments in order by matching overlapping regions
dNTP vs ddNTP
dNTPs are nucleotides that are building blocks of DNA.
ddNTPs are nucleotides that are used in the Sanger sequencing method.
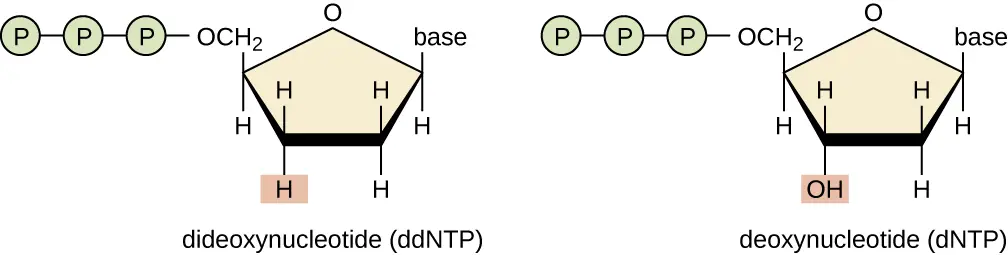
What is the name used for ddNTP used in DNA sequencing because they prevent further elongation of the DNA molecule when they are inserted into the molecule?
Terminator bases
Sequencing DNA Full Process:
The DNA for sequencing is mixed with solutions. The solutions are:
Primers
DNA nucleotides
DNA polymerase
A Terminator base (A, G, C, or T)
Place the mixture in a thermal cycler. This rapidly changes temperature at programmed intervals in repeated cycles.
At 90C, the double stranded DNA separates into 2 single strands
At 50C, the primers anneal to the DNA strand
At 60C, DNA polymerase adds nucleotides with complementary bases to the single strand DNA template. This build up new DNA strands.
Each time a terminator base is incorporated (instead of a normal nucleotide), the synthesis of the DNA is terminated as no more bases can be added.
Terminating bases are present in lower amounts and are added at random, resulting in many DNA fragments of different lengths depending on where the terminating bases were added.
After many cycles, all possible DNA chains will be produced. The reaction will have stopped at every base.
The DNA fragments are then separated by length
Terminator bases have fluorescent markers.
These markers are used to identify the final base on each fragment.
Lasers detect the different colours, and thus the order of the sequence
The order of bases in the capillary tubes shows the sequence of the new complementary DNA strand that was made.
From that, we can figure out the original DNA sequence.
What's a way to separate fragments?
Gel electrophoresis
Electrophoresis Process
Pipette the 4 solutions containing the 4 different terminator bases into the different wells in the agarose gel.
Pass a current through the electrophoresis plate from negative to positive.
DNA has a slightly negative charge so will be repelled by the cathode (-) and attracted to the anode (+)
Use an alkaline solution, as the solution carries charge to separate fragments.
Smaller fragments will travel further as they have less mass and therefore less resistance in the gel
Only run for a set amount of time, or else all the DNA will end up at the anode end.
Stain the gel then photograph the gel
2 ways to see the DNA after Gel Electrophoresis
Southern blotting
Radioactive/fluorescent probes
Southern blotting
Using radioactive DNA probes and x-rays.
It transfers fragments to a membrane.
Radioactive/fluorescent probes
These visualise bands/patterns
Difference between Electrophoresis and TLC
TLC separates by relative solubility
Electrophoresis separates by size
TLC separates non-charged particles
Electrophoresis only separates charged particles
TLC doesn’t use a buffer solution
Electrophoresis uses a buffer solution
TLC uses dyes
Electrophoresis uses radioactive / fluorescent tags
Faster techniques for DNA sequencing
High throughput sequencing
Next generation sequencing
Shotgun sequencing
Whole genome sequencing
Pyrosequencing
Massive parallel sequencing
Uses of DNA profiling
Paternity tests
Forensics
Assessing disease risk
Classification/species identification
Introns
Non-coding regions of the DNA
Explain why some regions of DNA can be described as non-coding
These regions are not present in mature mRNA and are not translated
Suggest why non coding regions of DNA show more variation
They aren’t selected against. They don’t affect survival.
Why do we use introns for genetic profiles?
- In most people, the genome is very similar
- Using coding sequences of DNA would not provide unique profiles
- Introns contain repeating sequences
STRs
Short tandem repeats
2-4 base pairs repeated 5-15 times
VNTRs
Variable number tandem repeats
20-50 base pairs repeated from 50 to several 100 times
What is the name for the sequence of bases that is repeated in Short tandem repeats (STRs) and variable number tandem repeats (VNTRs)?
The core sequence
DNA Profiling Process
Extract the DNA
Amplify the DNA fragment with PCR
Cut the DNA with restriction enzymes
Separate DNA fragments using electrophoresis. The DNA will be separated based on their mass.
Transfer fragments to paper
Apply a radioactive probe.
Use x-rays to view the position of DNA fragments
As a result, you create genetic profiles, which can be used in paternity tests, forensics, and for analysis of disease risk.
What are restriction endonucleases?
Restriction enzymes
They are enzymes used to cut the desired gene from DNA.
What can the desired gene also be called?
cDNA
Genetic Engineering Process
Restriction endonucleases are used to cut the desired gene from DNA. This creates sticky ends, which makes it easier for the gene to be inserted elsewhere.
The plasmid in this process is a circular piece of DNA used as a vector
A plasmid is cut using the same restriction enzymes.
This produces complementary sticky ends that match the gene.
The desired gene (cDNA) is inserted into the plasmid.
The complementary base pairs anneals using the enzyme DNA ligase.
It forms recombinant DNA.
A marker is often included with the desired gene. This helps identify which cells have successfully taken up the new DNA. Examples of a marker include a fluorescent marker, and a gene for antibiotic resistance
The recombinant DNA is inserted into the host cell using electroporation.
An electric shock makes the cell surface membrane temporarily porous, so the plasmids can enter the host cell.
The host cell divides by mitosis, reproducing the recombinant DNA too and producing the desired protein.
Finally, scientists test the cells to check if the gene was successfully taken up.
If a fluorescent marker was used, they check for fluorescence
If an antibiotic resistance marker was used, they grow the cells in antibiotics. Only the ones with the gene will survive.
What’s electrofusion?
Electrofusion combines 2 different cells.
1 cell with the desired gene, 1 host cell.
The cells are placed next to each other and given an electric pulse.
The electric shock will cause their membrane to temporarily break, allowing them to fuse into 1 hybrid cell.
The new hybrid cell contains genetic material from both original cells.
This hybrid can be grown into a GM organism.
Alternative methods to extract the desired gene
Using reverse transcriptase
Reverse transcriptase
You start with the mRNA (rather than the desired DNA) and you reverse it using reverse transcriptase to return to the cDNA (complementary DNA).
cDNA can now be inserted into a plasmid just like a normal gene.
What is recombinant DNA?
DNA combined from 2 sources
Vectors for Plants
Plasmid of Agrobacterium tumefaciens
Vectors for Bacteria
BAC (Bacteria artificial chromosome), virus
Vectors for Animals
BAC, plasmids, virus
Somatic gene therapy in cystic fibrosis
A harmless virus is genetically modified to carry a normal copy of the CFTR gene.
The virus is dripped into the lungs through a thin tube inserted into the nose.
Once inside the lungs, the virus enters the lung cells and delivers the healthy CFTR gene into the lung cell’s nucleus, where it becomes part of the cell’s DNA.
As the lung cells divide, they carry the normal gene, allowing them to produce functional CFTR protein, helping to relieve symptoms of cystic fibrosis.