Techn. Done - MCB104 - Lecture 17: Developmental Genetics & Personalized Genomics
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
45 Terms
What is Developmental genetics?
The study of genetic control of organismal development
Many genes that control development are highly conserved throughout evolution
Genes involved in key developmental decisions often occur in hierarchies
Ex: How does single cell of fertilized egg differentiate into hundreds of cell types?
How is all life related?
Cells of many eukaryotes have structural features in common: for example nuclei and mitochondria
Metabolic pathways are virtually identical in all organisms
Almost all cells use same genetic code
Many homologous proteins have highly conserved amino acid sequences
Means information acquired from a model organism can often be applied to other organisms
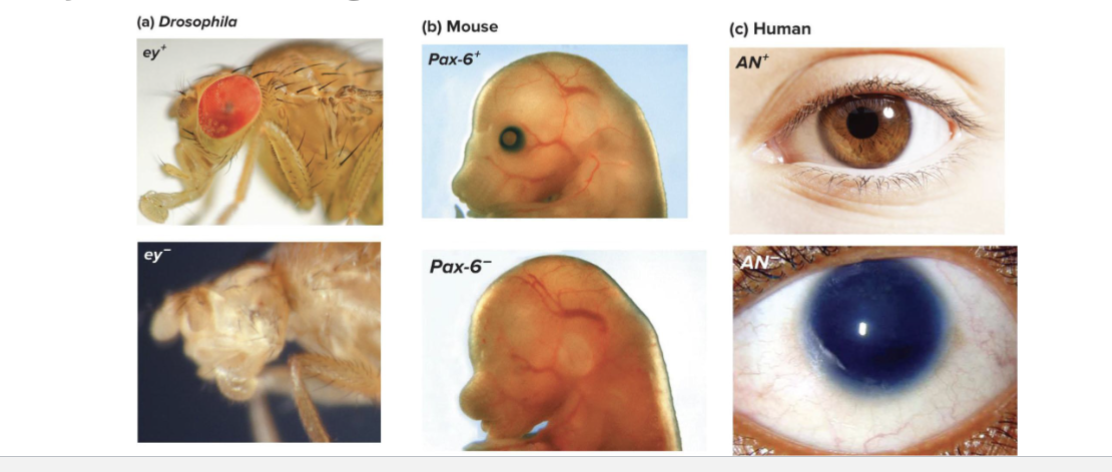
What does the Pax6 gene do?
Drosophilia, mice, and humans are mutant alleles of the eyeless/Pax-6/AN Gene
What are some different developmental schemes?
Disparate strategies are sometimes used by different organisms to accomplish the same developmental goal
Ex: cell fate in 2 cell embryos of C. elegans and humans
Mosaic determination in C. Elegans
Each cell has been assigned a developmental fate
Abnormal development if one cell is removed
Regulative determination in humans
Cells can alter their development fates according to environment
Twins result if 2 cells are separated
What’s the C. elegans lineage?
The location of the sperm fusing with the oocyte determined Anterior/Posterior polarity
The first cell division produces an anterior AB cell and posterior P1 cell
Each division has a different fate based on neighboring cells
Invariant lineage for all 959 somatic cells in hermaphrodite (1031 in males)
Screen Example: Drosophila Eye Development
Eye development is easy to study in flies
Visible, non-lethal
Ommatidium: Each optical unit of compound eye
In Drososphila there are 8 photoreceptors per ommatidium
How do you primary screen for an ommatidium defects
Mutagenized flies
Identified mutants where ommatidium lack 7th photoreceptor
Performed complementation tests and found 2 complementation groups
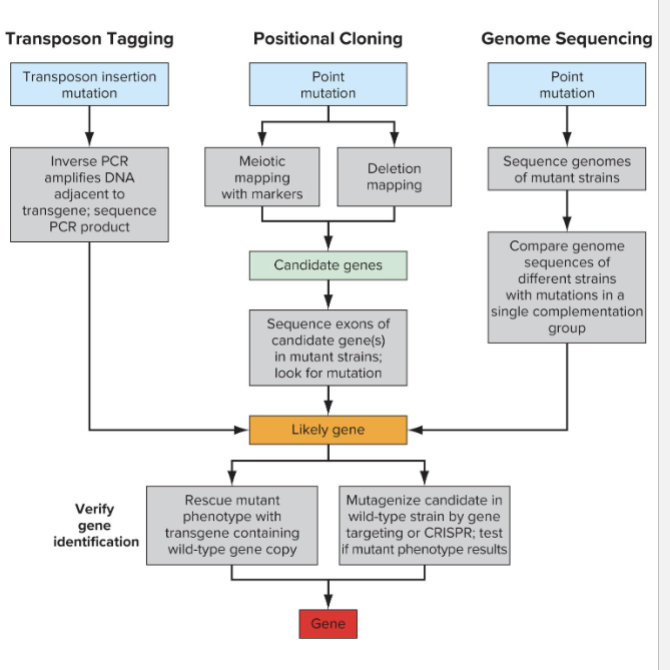
How do you identify mutations?
Mutagenesis method determines how you identify mutations
Transposon tagging allows inverse PCR to sequence flanking sequence
Positional Cloning narrows location on chromosome
Typically sequence candidate genes to validate
Genome sequencing identifies all polymorphisms
Compare sequences within complementation groups to find shared mutated genes
Verifying gene requires you either
Rescue with wild-type transgene
Recapitulate phenotype by introducing mutation into wild type strain
What is sevenless and bride of sevenless
The 2 genes identified in the screen for R7 loss are sev and boss
Structure gives clue for function
sev is an RTK
boss is an orphan GPCR (no known ligand)
boss is the ligand that activates sev activating a signaling cascade
Secondary Screen - What are Pleiotropy and Redundancy?
Some components of genetic pathways are missed in primary genetic screens
Pleiotropy: genes may be involved in several developmental pathways, including viability
Redundancy: 2 or more genes may perform the same function, a mutation in one will NOT result in a mutant phenotype
Secondary Screen- Why use Modifier Screen may identify pleitropic genes
Organism used for mutagenesis has a hypomorphic mutation that results in a sensitized background
Heterozygous mutations in a pleiotropic gene may modify phenotype caused by hypomorphic mutation
What’s a Modifier Screen for Pleiotropy?
Using a hypomorph, can screen for enhancing and suppressing mutations
Hypomorph is more sensitive to mutations than wild-type
What are transgenes for sensitized backgrounds
Can use transgenes to generate gain of function mutations
With appropriate regulatory sequences, can be limited to specific cells
Overexpression of Ras from transgene results in slightly rough eye
Additional mutations that enhance or suppress the phenotype were identified
What is involved in the Sevenless pathway?
DOS: daughter of sevenless
Not shown, scaffold protein
DRK: Downstream of Receptor Kinase
Enhancer of sevenless
SOS: Son of Sevenless (if hypomorph of SOS, it would be fewer R7, if knock it down, pathway lowers further, enhancement of SEV phenotype, but it wouldn’t affect the Ras, it’s always open)
Gas-GEF
Ras Pathway:
Ras: Monomeric GTPase
Res-GAP: Ras GTPase activating protein
RAF: MAPKKK
MEK: MAPKK
MAPK: Mitogen activated protein kinase
TFs
PNT: pointed, activator of R7 genes
Yan: anterior open, Antagonist of R7 genes
Show the pathway of Sevenless

What are some key points of the sevenless pathway?
Can you find:
Monomeric GTPase regulation
Kinase Cascade
Positive Regulation
Negative Regulation
Double Negative Regulation
For each protein can you predict whether it was identified as an enhancer or suppressor of
sevenless hypomorph
Ras gain of function
How can you do systematic screening of mutants
Mutagens alter genome at random, making traditional mutagenesis screens inefficient and incomplete
For model organisms, centralized centers maintain thousands of stocks, each with specific mutation
Many model organism have consortia whose goal is to generate knockouts of every ORF
Can be combined with nonmutagenic methods like RNAi or CRISPRi
Knockdown function in screen, acquire existing null mutant, assess phenotype
How can you analyze Developmental Pathways
Characterize action of each gene in pathway
Location and timing of gene expression
During development, where and when is mRNA found
Location of protein product
During development, where and when is protein found?
Developmental phenotype
What cells or tissues are affected by loss of function
How can you track RNA expression with RNA in situ hybridization
RNA in situ hybridization
Label cDNA of gene of interest
Use as probe to detect mRNA in organism
If Fluorescent probe, called Fluorescent in situ hybridization (FISH)
How can you track RNA expression with RNA seq
Determine sequences of all mRNAs in cell
Frequency of sequence relates to abundance in cell
How can you track protein localization?
Immunostraining
Use antibodies against specific protein
Specimen must be fixed
Translational reporter
Directly tag protein
Need to be certain tag doesn’t disrupt protein
How can you determine required location and timing? Example recombinant system?
Genetic mosaics - individuals composed of cells of more than one genotype
FLP/FRT recombination system
Similar to Cre/LoxP
Found naturally in yeast
FLP protein catalyzes reciprocal crossing-over at DNA targets called FRTs
FLP can be expressed from a transgene in certain cells
FLP originally not thermostable for mammals, but enhanced FLP (FLPe) is functional
What is FLP for Mosaics?
Used FLP FRT to induce mitotic recombination in dihybrid for gene of interest and linked marker gene
Cells lacking marker gene and lack gene of interest
Can characterize which cells require target gene to have wt phenotype
In sev this is R7
In boss this is R8
How can you determine timing
Temperature sensitive alleles can be activated/inactivated by changing temperature
Some other methods:
Chemical agonist/antagonists
Photoinactivation with tagged genes
How can you determine Order
If loss or overactivity of gene A function affects expression of gene B and not the converse, then gene A functions upstream of gene B in a genetic pathway
Ex:
Expression of prospero is activated by Ras, but Ras isn’t activated by prospero
Therefore, Ras is upstream of prospero
How can you determine Epistasis, Developmental Pathway Edition
Start with mutations in Gene A and B
Phenotype of double mutant resembles one of single mutants
Usually defines earlier acting step in pathway
Useful in investigating switch/regulation pathways (on/off)
Required Conditions
Phenotypes of single mutants must be different
Mutant alleles must be null or constitutive
Epistasis and Sevenless - describe Ras?
Ras gain of function transgene masks sev- phenotype
Ras is downstream of sev
Describe eyeless/Pax6
Identified in mutagenesis screen
Phenotype - no eyes
In humans, mice and flies, loss of Pax6 leads to defects in eye formation
How do Expression of Pax6 work
In situ hybridization probe for Pax6 mRNA in human fetal tissues
Expression is in areas that will eventually, produce eyes
Suggests potential conserved role
How can you identify expression patterns of regulatory sequences?
Isolate segments of DNA around gene of interest
Insert segments of vector containing minimal promoter and reporter gene
Insert constructs into organism
Observe expression pattern of reporter
Often done by sequentially narrowing down fragment size
Sometimes identified via CHIP-seq data
How does Regulation of Pax6 occur?
Many enhancers regulate expression of Pax6
Note retina enhancer is intron
Many of transcription factors targeting Pax6 enhancers are homeobox genes
What are Pax6 Regulatory Targets
Can use CHIP-seq to identify specfic enhancers that Pax6 binds
Different Tissues have different TFs, leading to different expression patterns
What is GAL4/UAS for Ectopic Expression
GAL4 is a yeast transcription factor and very strong activator of gene expression
Upstream Activation Sequence (UAS) is the enhancer sequence GAL4 binds
Conditional Expression lines can be made using 2 constructs
GAL4 with a tissue specific regulatory sequence
Desired gene with UAS
Modular system can simplify experimental setups
Is Pax6 sufficient for eye formation?
Yes
Expressing Pax6 ectopically using different Gal4 constructs to activate UAS driven Pax6 transcription leads to formation of eyes
What is Evolutionary connection and Pax6
Cavefish often lose vision over evolutionary time
One mechanism is loss of Pax6 expression in their optic buds
What is personalized Genomics
Genetics can influence a wide range of medically relevant characteristics
Disease risk
Drug Responses
Metabolic Processes
Immune Function
Personalized Genomics is the study of individual’s genome in order to make predictions about range of traits
Often applied to medical purposes
What is Prenatal and Neonatal Genetic Testing
Most widespread example of personalized genetics, samples are taken from embryos/fetuses and checked for various genetic diseases
includes
testing for various aneuplodies and chromosomal rearrangements
identifying known mendelian disease alleles
Describe Cancer Genomics/Precision Oncology
Variants of specific genes are associated with increased risks of cancer
Sequencing tumors can help make predictions as to tumor behavior
Combining individual and tumor WGS can guide therapeutic strategies
What Infectious Disease Genomics
Sequencing can be used to identify specific variants of an infectious agent, serving as diagnostic tool
Specific genetic variants influence susceptibility to different pathogens
Heterozygotes for the sick cell allele are more resistant to malaria
Homozygotes for the CCR5 -Δ32 allele to resist HIV infection but are most susceptible to west wile virus
Identifying those variants allows patients to better estimate risks
What is Pharmacogenomics
Pharmacogenomics is the study of gene/drug interactions
Drug metabolism includes various functions including
Uptake
Release
Breakdown
Products
Each of these functions involves various genes
Alterations in function of these genes can alter effectiveness, optimal dosage, and side effects of drugs
Ideally, one could sequence a patient’s genome and use the results to identify which drugs and at which dosage would be the best
What is Warfarin
Widely prescribed anticoagulant drug used to prevent blood clotting related diseases in patients with deep vein thrombosis, atrial fibrillation, recurrent strokes or heart valve prosthesis
High inter-individual variance in optimal dosing combined with narrow therapeutic range making prescribing it challenging
Dose too low: blood clotting
Dose too high: internal bleeding
Need to identify sources of variation to predict ideal dosage
What is Warfarin Mechanism of Action
Warfarin is an inhibitor of vitamin K epoxide reductase complex subunit 1 (VKORC1)
Inhibition of VKORC1 reduces amount of Vitamin K available as cofactor of clotting proteins
Variation of VKORC1 accounts for ~30% of variation in warfarin dose
What are Warfarin Pharmacokinetics
Warfarin is a mixture of R and S stereoisomers
S stereoisomer is primarily metabolized via CYP2C9
Variation of CYP2C9 and VKORC1 together account for ~40% of variant in dose
Adding age, weight, height, interacting drugs and symptoms only predicts about ~55%
How can you interpret data correctly
Studying effects of specific variants in human genome is extremely challenging
Literature full of associations based on
Very low N, including case reports
Single studies failed to replicate
In vitro/molecular/functional assays
Generally, put more trust in connections that have been demonstrated in multiple studies