Chapter 2: Gene Expression and Epigenetics Molecular Diagnostics Lela Buckingham
1/134
Earn XP
Description and Tags
https://quizlet.com/697841530/chapter-2-gene-expression-and-epigenetics-molecular-diagnostics-lela-buckingham-flash-cards/
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
135 Terms
What is transcription
A way of gene expression. Done by copying 1 strand of DNA into RNA via RNA polymerase. Cells with the same genotype can have vastly different phenotypes due to structural changes in DNA from methylation
Which stage of the cell cycle does transcription/translation occur
interphase
RNA polymerase I and III codes for
noncoding RNA
RNA polymerase II codes for
mRNA
RNA Polymerase: E coli RNA Polymerase II
Uses DNA to produce mRNA
RNA Polymerase: RNA Poly I
Uses DNA to produce rRNA
RNA Polymerase: RNA Poly II
Uses DNA to produce mRNA
RNA Polymerase: RNA Poly III
Uses DNA to produce tRNA and snRNA
RNA Polymerase: mitoRNA polymerase
Uses DNA to produce mRNA in the mitochrondria
RNA Polymerase :mammalian DNA poly a
Uses DNA to produce primers
RNA Polymerase: Dengue Virus Polymerase and HCV RNA Polymerase
Uses RNA to produce its own viral genome
RNA Polymerase: PolyA polymerase
Produces PolyA tails without a template
How is information in DNA utilized
It has be to transcribed and then translated into protein
Cellular activity and location of RNA Polymerase I in eukaryotes
Found in the nucleolus, Produces 18S, 58S, and 28S rRNA insensitive to alpha-amanitin
Cellular activity and location of RNA Polymerase II in eukaryotes
Found in the nucleus. Produces mRNA and snRNA and Inhibited by alpha-amanitin
Cellular activity and location of RNA Polymerase III in eukaryotes
Found in the nucleus. Produces tRNA and 5s rRNA. Inhibited by high concentration of alpha-amanitin
Transcription initiation
RNA and its accessory proteins assemble at start site at DNA (Promoter). RNA has a lot more transcription promoter sites when compared to DNA even though DNA and RNA catalyze basically the same reaction. Also, RNA polymerases work with different proteins to find/bind DNA for transcription preparation.
Transcription elongation
RNA is synthesized using the base sequence of one strand of the double helix (anti-sense strand). RNA Polymerase move slower than DNA polymerase but DON'T REQUIRE PRIMING. 1st ribonucleoside triphosphate retains all of the phosphate groups but the subsequent ones only retain alpha PO4. 5'5' covalent cap attached to methylated guanine residue after initiation of RNA synthesis. The Cap protects the end of the growing pre-mRNA, facilitates ribosome binding and efficient translation
Transcription Termination In prokaryotes
increase in RNA synthesis occurs when protein products are low, increase levels of gene product induce termination of synthesis. Done by interaction of RNA polymerase w/ DNA template or Rho interaction. Rho interaction starts at the G-C rich area hairpin and stops at the A-T rich area.
RNA Polymerase I Termination factors
Sal box and TTF 1
RNA Polymerase II Termination factors
Alternate polyadylation sites
RNA Polymerase III Termination factors
adenine residues/TF
Transcription Termination In eukaryotes
mRNA synthesis (catalyzed by Pol II) Proceeds until it reaches a polyA site. The just formed mRNA is released by an endonuclease and subsequent RNA is degraded 5' to 3' towards RNA polymerase until it reaches PolyA site where transcription stops and PolyAdenate Polymerase adds PolyA tail of 20-200 Adenine. This is important for stability, localization, and translation
lac operon
a gene system whose operator gene and three structural genes control lactose metabolism in E. coli
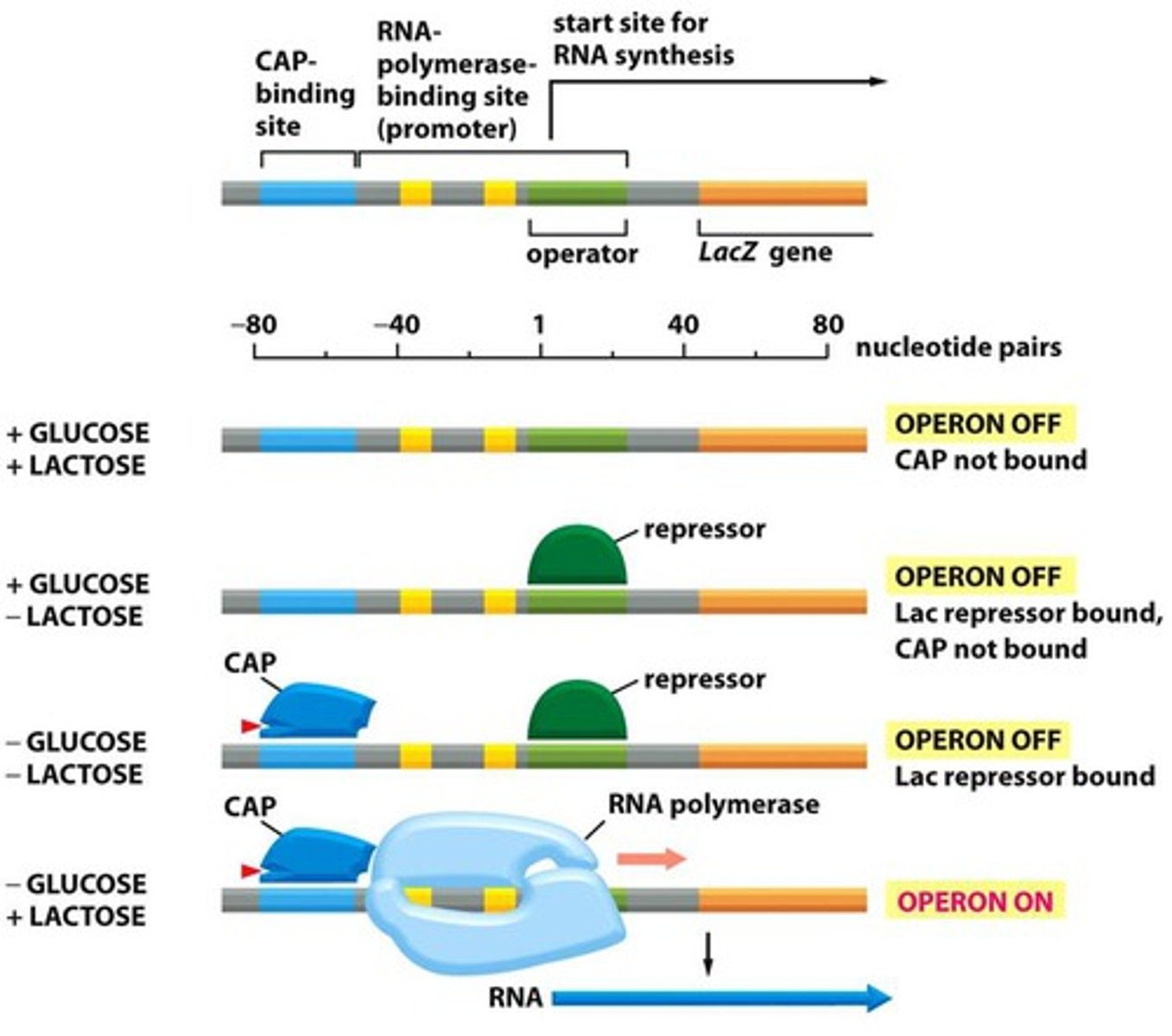
Regulation of Transcription
Gene expression is a key determinant of phenotype. Can be constant or have tight regulation. Stoichiometric balance with specific combinations of proteins are crucial for all differentiation/development protein availability; Protein availability and function are controlled via transcription, translation, and protein modification
constitutive
always on, continual use within the cell
Regulation of mRNA synthesis at initiation is done by
Cis factors and Trans factors
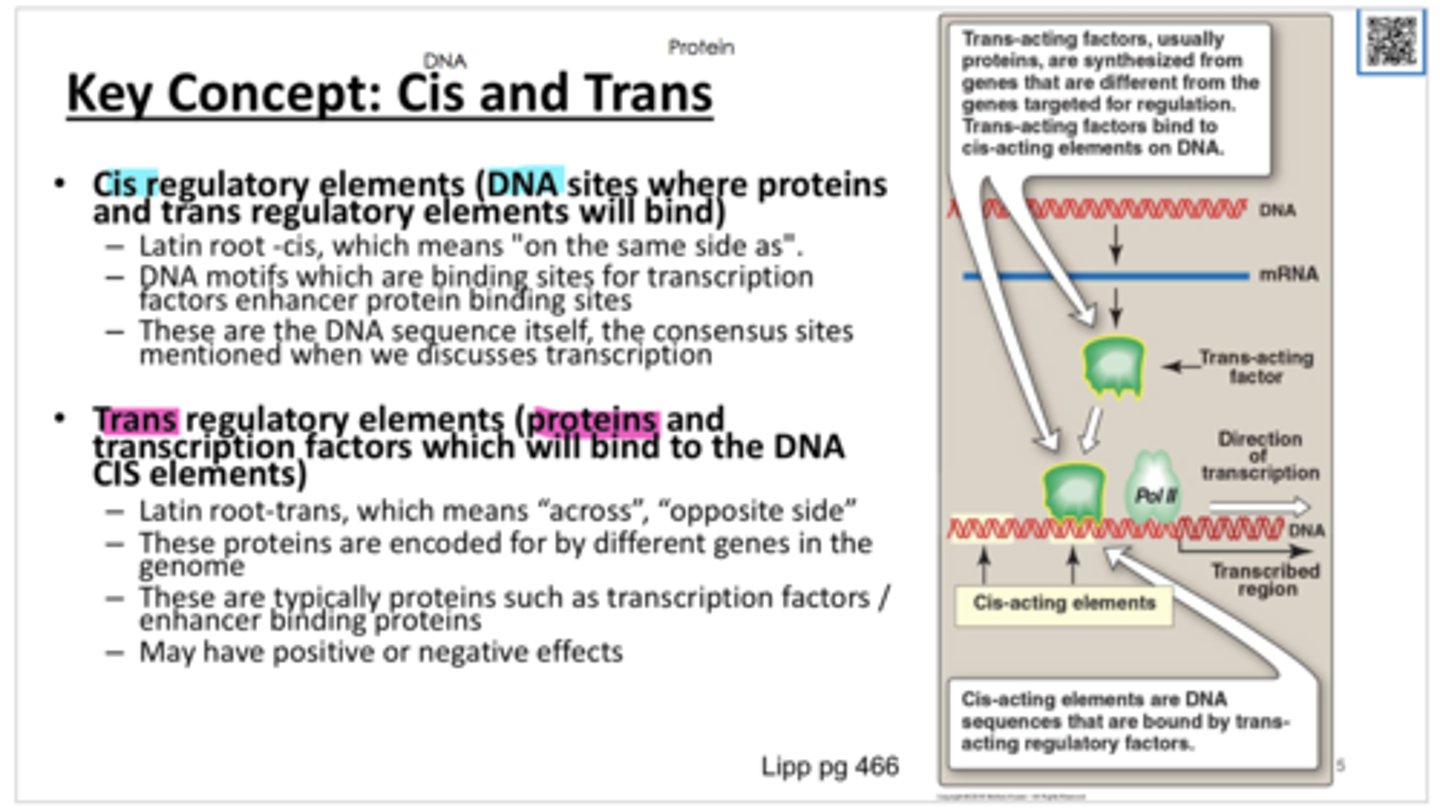
cis factors
DNA sequences that mark places on DNA involved in initiation and control of RNA synthesis
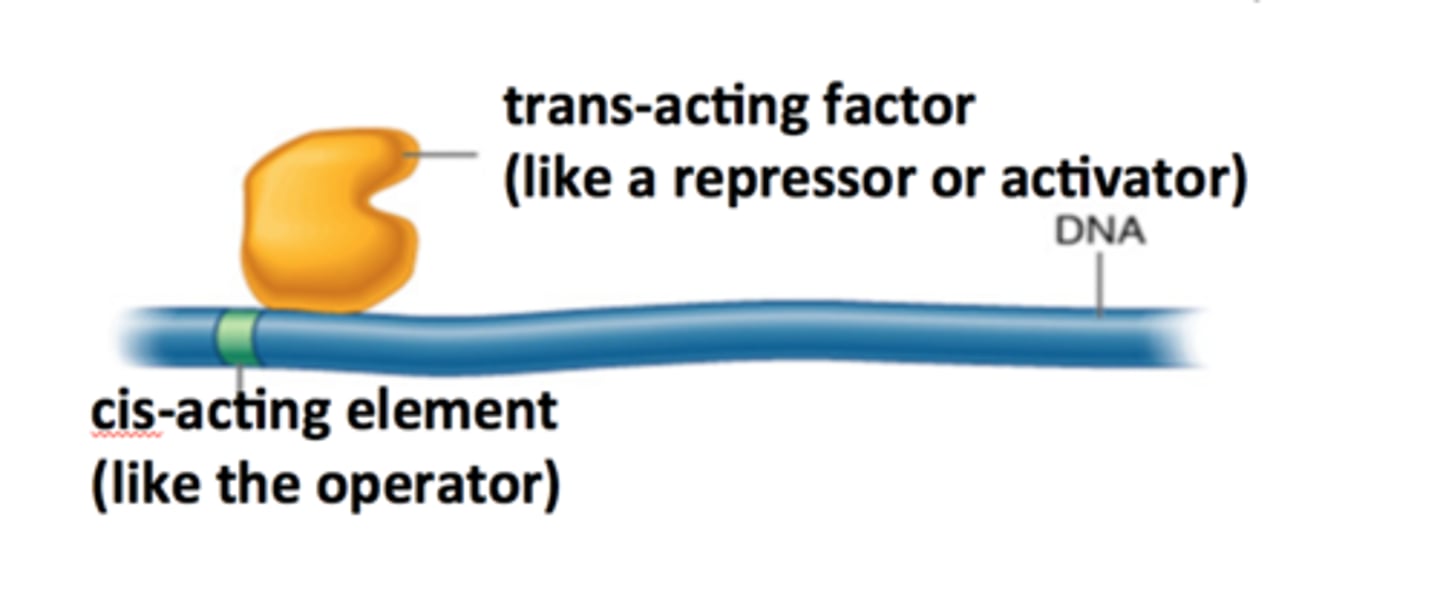
Trans factors
distant, diffusible, proteins (activators and repressors) that interact with the cis elements
operon
A unit of genetic function common in bacteria and phages, consisting of coordinately regulated clusters of genes with related functions and small genomes. Transcribed on one mRNA and subsequently separated into individual proteins
Operons are controlled by binding regulatory trans factors to cis sequences preceding the structural genes
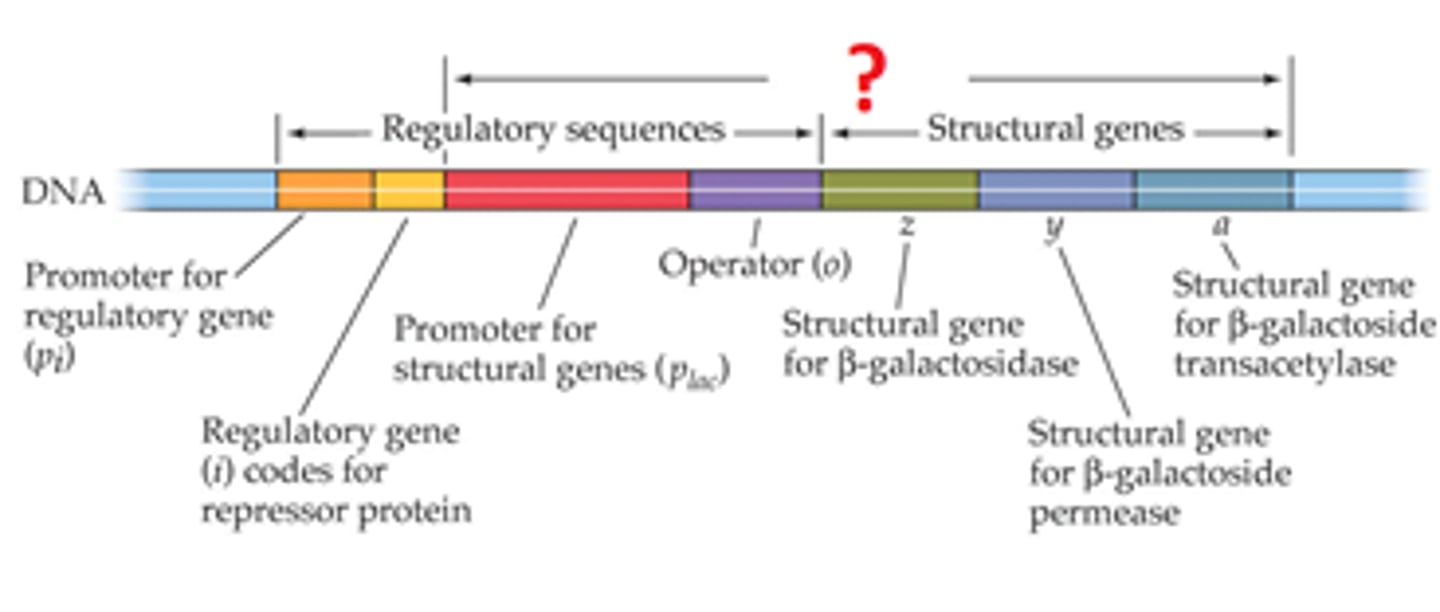
Enzyme Repression
type of negative control where a co-repressor must bind to a repressor in order to turn off transcription
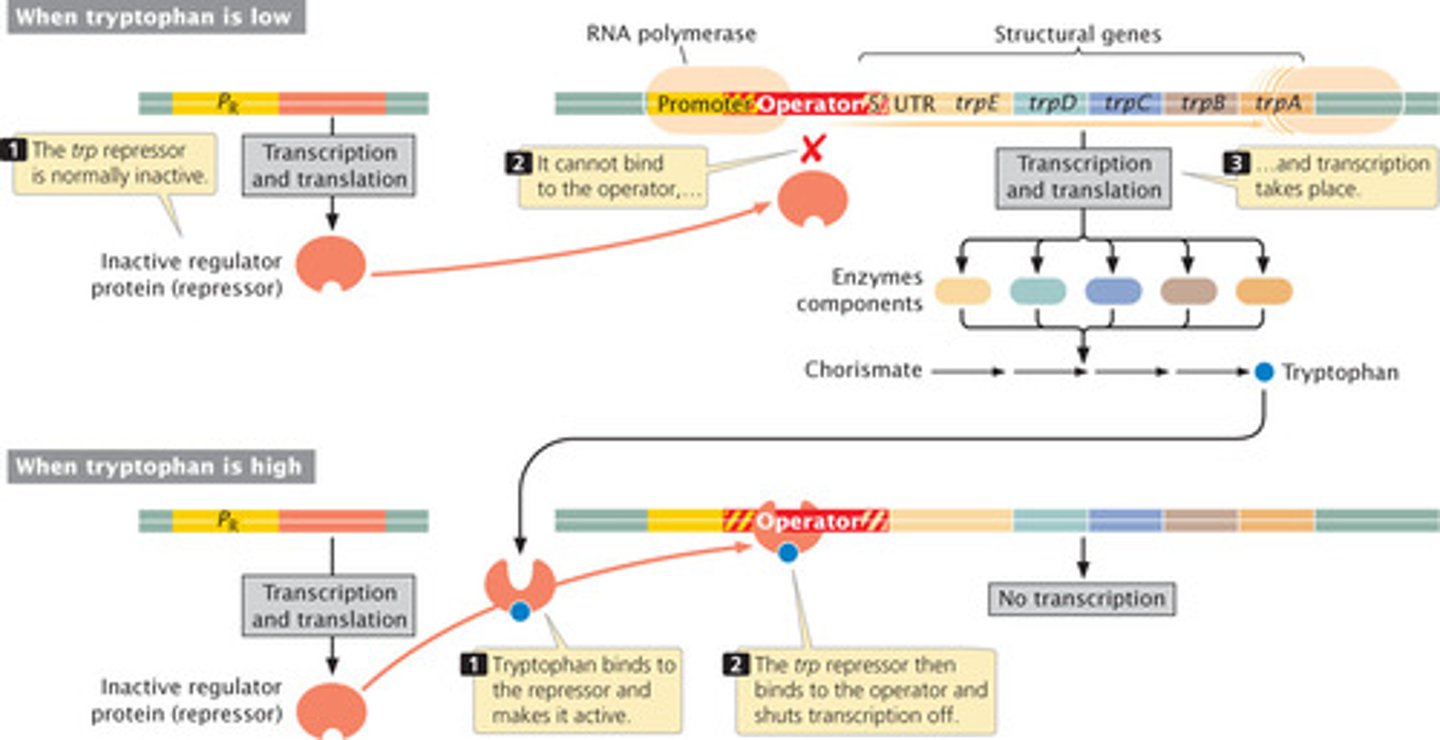
enzyme induction
inducer will bind to the repressor to leave the operator free to turn on expression for transcription
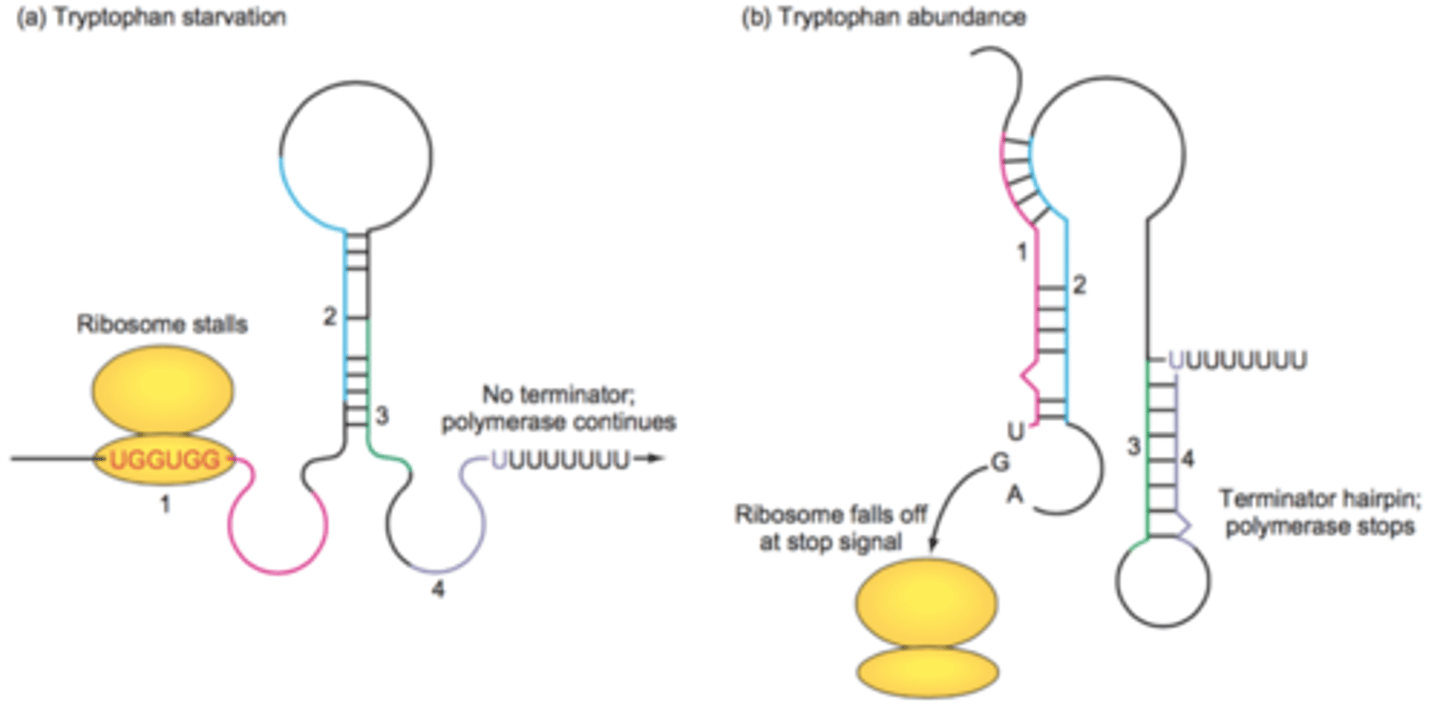
Enzyme attenuation
regulation through formation of stems and loops in the RNA transcript by intra-strand H bonding of complementary bases
Regulates expression at the translational level based on amount of enzyme present
Anti-termination- will proceed with translation
Termination- high levels will get stopped by hairpin to stop translation
cis regulatory elements: Prokaryotes
Their regulatory elements are close to the structural gene they control near the promoter
Cis regulatory elements: Eukaryotes
Distal elements can be far away from the genes they control, proximal elements cam be located in or around the genes. Elements can be behind the genes. more complex.
If Eukaryotes don't have operons, how are genes expressed
Synchronous expression via combination control. Similar patterns cause similar cis elements to respond simultaneously to specific combinations of controlling trans factors
post-transcriptional regulation: Prokaryotes
only changes through transcription process can cause changes in gene expression and transcription and translation is concurrent. RNA stability can be affected by secondary structure and polyadenylation
Post-transcriptional regulation: Eukaryotes
Trickier; Regulated by RNA processing steps and Alternate splicing. Alternate splicing with protein factors are responsible for tissue and development specific gene expression patterns
Circular RNA
Controls gene expression. Endogenous byproducts of RNA splicing in protein coded genes. Roles are: protein binding, activation of transcription, sub-cellular localization, stable. Implicated int tissue development, stress response, and cancer
Post Translational regulation
Peptides and proteins are modified with lipids, sugars, and other factors by cellular enzymes such as kinases, phosphatases, ubiquitinases, transferases, acetylases, methylases, and ligases
Gene expression is measured in the lab at the level of transcription and protein production
Epigenetics
Phenomenon that allows cells to take on different phenotypes without changing its genotype. Changes occur at the meiotic and mitotic level not encoded in genotype. Rapid and heritable due to environmental changes w/o changing DNA. Modified by chromatin and noncoding RNA
histone modification
adding chemical modifications to proteins called histones that are involved in packaging DNA. Chromatin nuclear DNA and associated proteins, compacted into nucleosomes in eukaryotes
Nucleosome
150 bp of DNA wrapped around 8 histone proteins: 2 Histone 2A, 2B, Histone 3, Histone 4. This modifies histone by modifying chromatin protein and transcription factors affecting gene production.
Occurs on specific amino acid and carboxy ends of histone protein sequences
What are ways that nucleosomes can modify histones to affect gene production
Methylation, phosphorylation, ubiquination, and acetylation.
How to notate the Histone Code
Nomenclature for Core Histone + Amino Acid Position (single letter code) + Modification '
Acylation of lysine at position 27 @ Histone H3= H3K27Ac
Histone H2A Acetylation Function
activation
Histone H2A phosphorylation function
mitosis/DNA repair Function depends on amino acid modified or combination of modifications present
Histone H2A Ubiqutylation function
stem cells
Histone H2B Acetylation Function
activation (Like H2A)
Histone H2B phosphorylation function
Apoptosis
Histone H2B Ubiqutylation function
Transcription
Histone H3 unmodified function
silencing
Histone H3 methylation function
silencing/activation Function depends on amino acid modified or combination of modifications present
Histone H3 Phosphorylation function
mitosis/activation Function depends on amino acid modified or combination of modifications present
H3 Histone Acetylation Function
Activation/histone positioning Function depends on amino acid modified or combination of modifications present
Histone H4 phosphorylation function
Activation
Histone H4 methylation function
silencing and activation Function depends on amino acid modified or combination of modifications present
H4 Histone Acetylation Function
DNA repair and histone positioning
What are histone deactylase inhibitors used for
hematological malignancies (cancers) and can be used as a therapeutic or diagnostic tool
Non-Polar Amino Acids
Alanine, Leucine, Isoleucine, Proline, Phenylalanine, Methionine, Tryptophan, Valine
polar amino acids
Serine, Threonine, Tyrosine, Asparagine, Cysteine, Glutamine, Glycine, Proline
Negatively Charged amino acids
aspartic acid, glutamic acid
Positively charged amino acids
lysine, arginine, histidine
alanine abbreviation and letter
Ala, A
Isoleucine abbreviation and letter
Ile, I
Leucine abbreviation and letter
Leu, L
Metionine abbreviation and letter
Met, M
Phenylalanine abbreviation and letter
Phe, F
Tryptophan abbreviation and letter
Trp, W
Valine abbreviation and letter
val, V
Asparagine abbreviation and letter
Asn, N
Cysteine abbreviation and letter
Cys, C
Glutamine abbreviation and letter
Gln, Q
Glycine abbreviation and letter
Gly, G
Proline abbreviation and letter
Pro, P
Serine abbreviation and letter
Ser, S
Threonine abbreviation and letter
Thr, T
Tyrosine abbreviation and letter
Tyr, Y
Aspartic Acid abbreviation and letter
Asp, D
Glutamic Acid abbreviation and letter
Glu, E
Arginine abbreviation and letter
Arg, R
Histidine abbreviation and letter
His, H
Lysine abbreviation and letter
Lys, K
DNA methylation is done with ________________ in prokaryotes/eukaryotes
epigenetic regulation
Where does epigenetic regulation occur in vertebrates
In CG rich areas or regions of more than 200 bp in length with an observed/expected ration of greater than 0.6. Found around: 1st exons, promoter regions and 3' end of genes
Imprinting
mainly done in the methylation of DNA stage. Gamete specific silencing of genes. Maintains balanced expression of genes in growth and embryonic development by selective methylation of homogenous genes. Examples where imprinting made a difference: mules vs hinnies, Angelmann vs Prader Willi Syndrome. This phenomenon may also be partially responsible for abnormal development of phenotypic characteristics of cloned animals because cloning bypasses gametogenesis
Mules and hinnies are both the resulting of hybridizing between a horse and a donkey. This shows the difference between
imprinting during oogenesis and spermatogenesis
Angelmann/Prader Willi
show genomic imprinting problems on chromsome 15
What are the four enzymes that methylate DNA
DNMT 1- maintenance enzyme for hemi-methylated DNA
DNMT 3a and 3b- de novo methylase that methylates un methylated DNA establishing newly methylated regions as seen in imprinting
DNMTL- non functional. May regulate 3a/3b by occupying DNA and protein sites.
DNMT
DNA methyltransferase
DNA methyltransferase 2 may modify
bases in tRNA and others
DNMT 1 and 3 may recognize
cytosine followed by guanine bases based on their symmetry. These enzymes are recruited to methylated areas by proteins that bind histones and hemi-methylated DNA
Transcriptional silencing
responsible for X chromosome silencing in females and position effects (Silencing genes when placed in heterochromic areas)
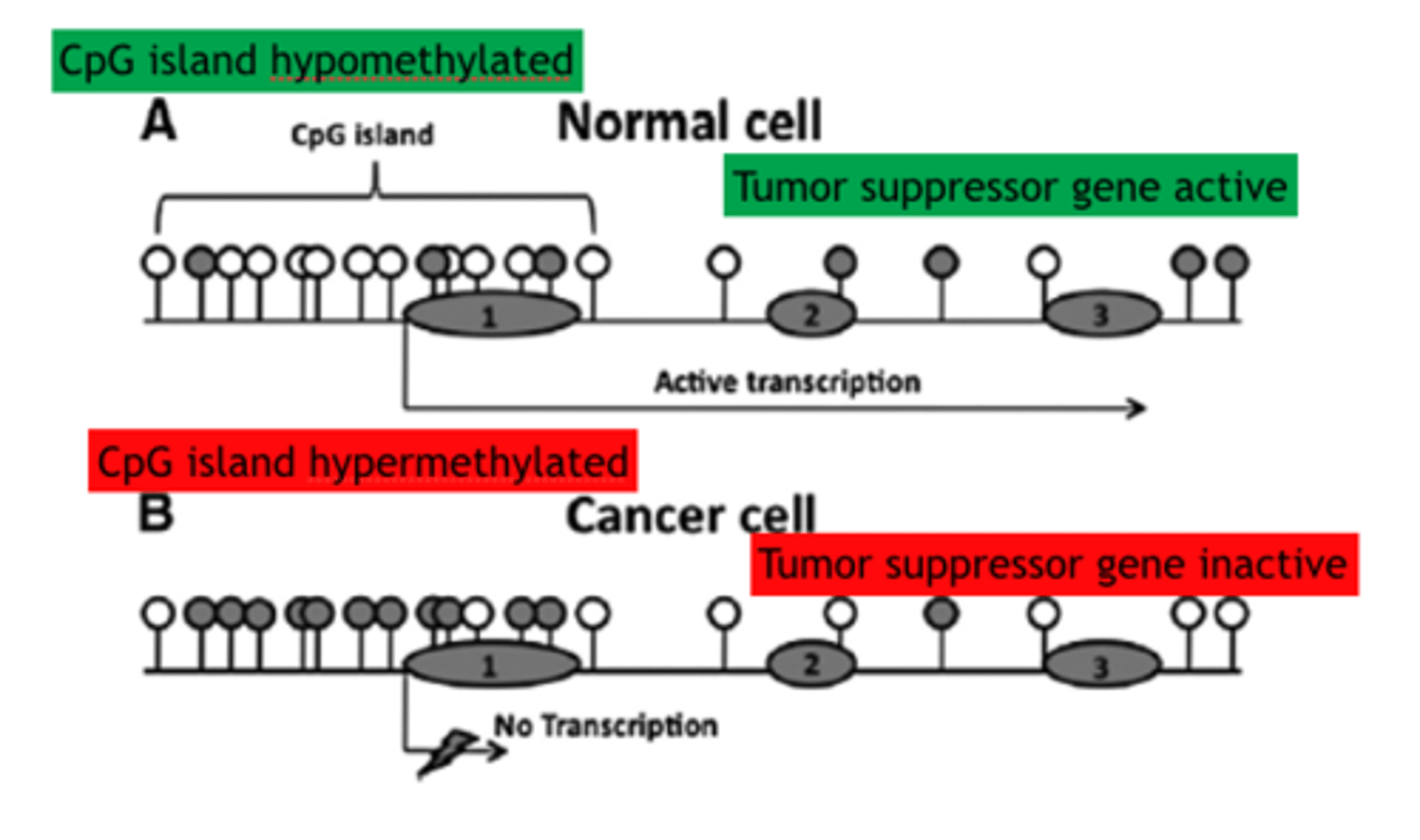
CpG islands
DNA regions rich in C residues adjacent to G residues. Especially abundant in promoters, these regions are where methylation of cytosine usually occurs.
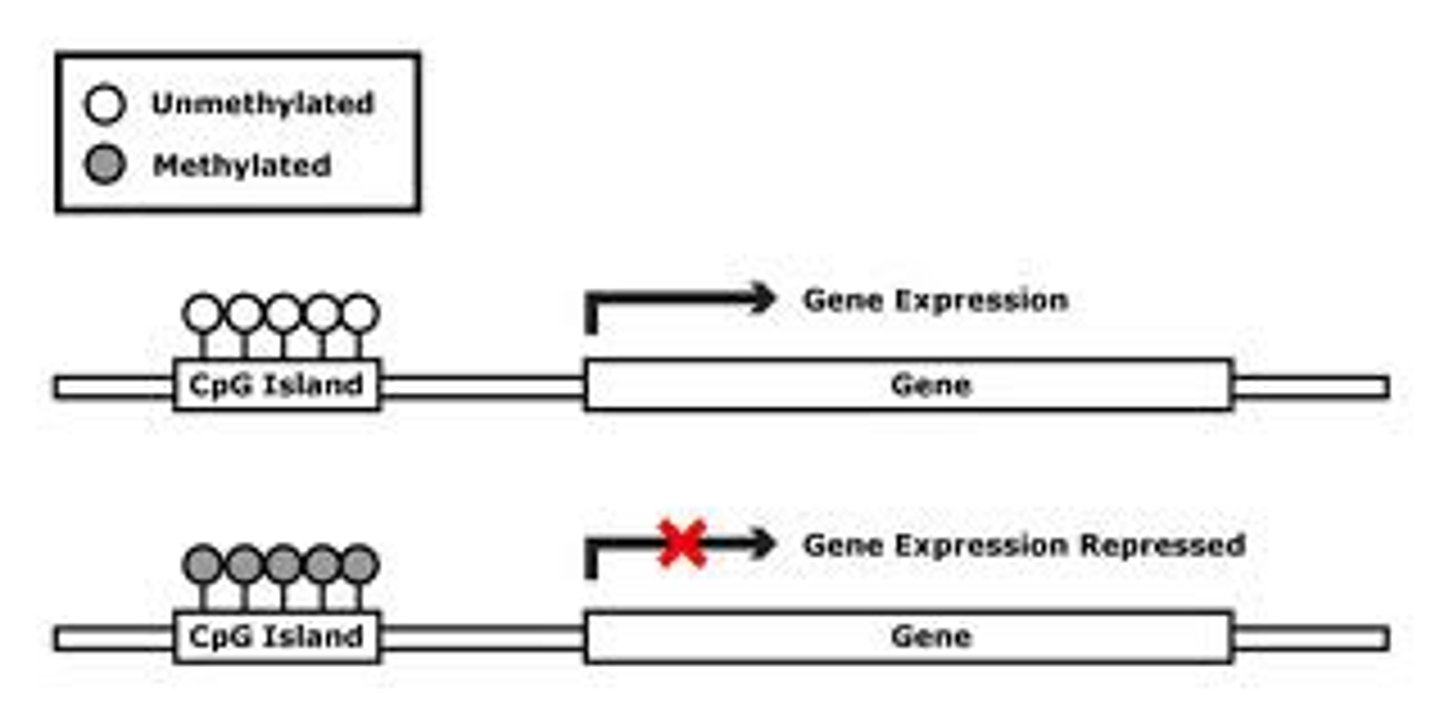
Normal CpG Islands
unmethylated or transiently methylated islands before active genes. Areas can also have heavier methylation before coding sequences
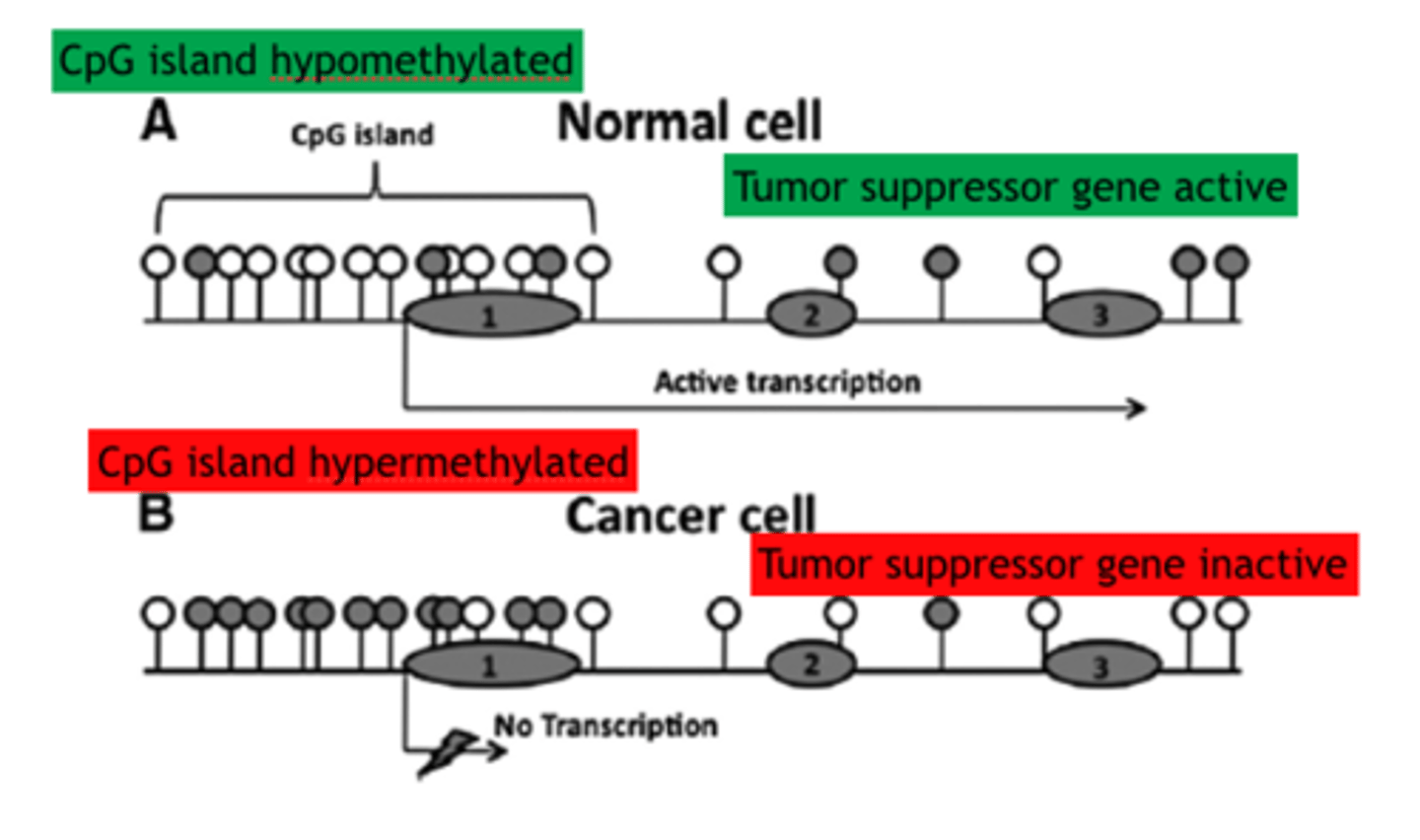
Abnormal in CpG islands
Colon and Brain cancer causing hypermethylation CpG island Methylator Phenotype (CIMP), Hypomethylation can also occur in tumor cells and if it occurs around long interspersed nuclear elements (LINES) it can cause genomic instability in cancers and chromosome alterations/breakages
DNA demethylation
The removal of the methyl group from methylated DNA enabling genes to become active so they can be transcribed
Ten-Eleven Translocation Enzymes (TETs)
enzyme which catalyzes demethylation through a series of oxidative reduction reactions. Major pathway of active demethylation in vertebrates and require a-ketogluterate to function
1.) 5-methylcytosine-->hydroxymethylcytosine
2.)hydroxymethylcytosine -> 5 formyl methyl cytosine
3.) 5 formyl methyl cytosine-> 5 carboxymethyl cytosine
4. Decarboxylase enzyme is added to restore 5 methyl cytosine
RNA methylation
occurs in tRNA, rRNA, mRNA, and ncRNA. Usually on N6-methyladenosine @ GAC or AAC. 25% have m6a around stop codons. Sometimes found in 5' untranslated regions of RNA.
This effects RNA stability, splicing, and translation
Methyltransferase enzymes RNA
METTL14 and METTL13