NUT 116 Lec 4 - Nutritional Phenotype
1/32
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
33 Terms
most chronic diseases are
complex/multiple gene disorders or polygenic diseases
clinical phenotype
susceptibility genes + environmental factors
diets do not…
benefit everyone the same
EX of heterogeneity of biologic responses to nutrition inputs
gave participants fat based meal and measured TAG
see huge variability in ppl
influenced by metabolic traits (weight, insulin resistance)
or other factors ( behavior, microbiome)
5 individuals, 5 diff responses
heterogeneity of responses can be related to
phenotypic differences
can be observed in lean vs obese state
bw
blood pressure/hypertension
insulin resistance
diabetes
phenotypic differences can influenced
metabolic response to nutrition
body size, adiposity
presence of disease or disease risk factor
microbiome
precision or personalized nutrition seek to understand phenotypic differences and better predict responses to
make tailored dietary reccs
variation in response is related to
biological and phenotypic complexity
biologic and phenotypic variability
complex interactions between multiple
gene x environment; gene x diet; diet x microbiome interactions
epigenetic changes
underlying pathology or disease risk
metabolic flexibility
ability of cells and tissues to switch fuel sources, alter gene expression, adapt to various stressors, magnitude of resposes
why so much variability in diff individuals’ responses to foods and diet patterns
health affected by complex, dynamic interactions between genetics, epigenetics, diet, environment and lifestyle, physio state and age
diet is heterogenous and adds complexity to diet x gene and die metabolism interactions
how monitor and study metabolic responses?
omics approach
omics approach
genomic
what can happen and what appears to be happening (transcriptomic)
based on DNA person has and RNA being produced
proteogenomic
what makes it happen
proteins being made based on active gene
metabolomic
what has happened and what is happening in metabolic pathways measured by circulating or excreted metabolites
types of gene-environmental interactions and effects on a health risk
risk allele present, health risk is greater even without environmental exposure
with environmental exposure, magnitude of health risk significantly greater than when risk allele present
even medium dose of environmental exposure greater effect when risk allele present
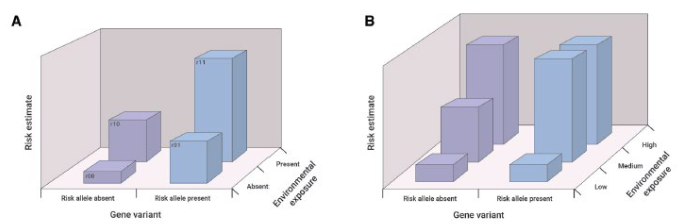
type of gene-environment interactions and effects on health risk
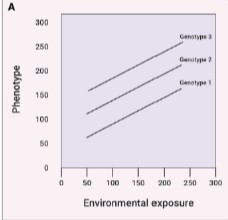
health risk associates with risk allele but all genotypes change in same way to exposure
slope of lines same/parallel
no gene x diet interaction
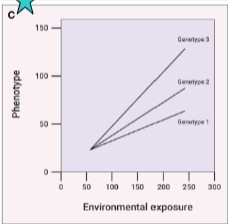
type of gene-environment interactions and effects on health risk
effect of diet somewhat similar on all alleles (same direction), but slope differs by genotype
diet exposure differentially affects genotype (slope of line changes)
gene x diet interaction present
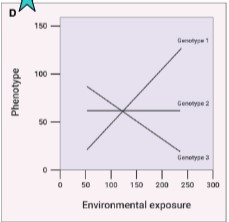
type of gene-environment interactions and effects on health risk
diet affects each allele of gene differently, differential effects of diet clearly seen by genotype
slope and direction of all lines vary
gene x diet interaction present
metabolomics
identification and quantification of metabolites in biologic stem
metabolites: low MW compounds, are reactants, intermediates, or products of biochemical rxn
measured in biologic fluids
tool and analytical apporach allows profile of biochem of individual to be described
links genotype with phenotype
assumes individual’s metabolic state reflects their health status
provides means to look at complex and integrated responses to diet and other exposures
nutritional metabolomics
marker of dietary intake
marker of metabolic effect
metabolomics as biomarker of dietary intake and diet composition
measure of diet exposure
reflects food components, their metabolites, and metabolites from gut microbiota
metabolomics uses in nutrition
assess validity of dietary assessment in observational trials
assess compliance during clinical trials
provide objective biomarkers for dietary intake
specific foods
dietary patterns and food groups
EX of metabolites originating from food (biomarkers of dietary intake)
food additives: sorbitol
FA
butter: myristic acid
walnut: a-linoleic acid
animal foods
chicken: anserine
red meats: acylcarnitine
plant foods
citrus: proline betaine
beverages
coffee: caffeic acid, ferulic acid, trigonelline
polyphenol-rich foods
olive oil: hydroxytyrosol
apples, choc: epicatechin
citrus: naringenin
grapes: tartrate
EX quantifying dietary intake based on metabolomics data
goal: quantify intake based on metabolites, RCT intake of OJ
metabolite proline betaine marker of OJ intake
predicted citrus intake from urine samples
metabolomics as biomarker of effect and associated health benefits
reflects endogenous intermediary pathways and metabolites
includes biomarkers of disease, disease risk, and health status
can describe metabolic phenotype, or metabotype
metabolomics uses
provide insight into mechanisms and pathways invovled in diet, disease relationships
provides ability to look at patterns of changes in response to diet, and physiological status
provides ability to distinguish between groups of individuals such as responders and non responders
EX: metabolomic signature associated with BMI and visceral adiposity
goal: examine relationships between BMI and 145 metabolites
goal: identify biomarkers of high vs low visceral fat area
future applications: metabolite markers of BMI and adiposity status for clinical use, investigations of change in biomarkers with dietary interventions, examine candidate biomarkers for causal pathway relationships with obesity related diseases
metabolomics applications
current uses and applications
predom research based
pilot studies, proof of concept
identification and validaiton of biomarkers of exposure and effect
diet-disease and diet-gut microbiota disease relationships
generate hypotheses
generating a lot of data
future use
identify nutritionally “high risk” patients
establish precision nutrition recc to prevent and manage disease
develop innovative education and counseling appraches based on personal phenotypic metabolomics info
metabotyping
grouping individuals based on similarities in metabolic characteristics and phenotype (metabotype)
consider factors such as diet, anthropometric measures, clinical parameters, metabolomics data, gut microbiota
metabotyping clinical usefulness
parameters to determine metabotype need to be easily measured and cost effective; and able to identify responders with associated health outcome and surrogate health biomarkers
personalized/precision nutrition
application of nutritional -omics, seeks to
understand factors that influence individual variability in response to nutrients
develop algorithms to predict response based on phenotype
make dietary reccs or interventions based on individual’s nutritional requirements, nutritional status, genotype, microbiome, metabotype, and phenotype
not one size fits all
goal of personalized nutrition
to preserve or increase health by using genetic, phenotypic, medical, nutrition, and other relevant info abt individuals to deliver more specific healthy eating guidance and other nutritional services
personalized nutrition - time is now
no longer theoretical
postprandial responses to same food vary between individuals
studies show personalized dietary interventions:
improve glycemic control and lipid profiles
enhance weight management and dietary adherence
commercial platforms use AI to tailor nutrition advice
multi-omics and machine learning enable prediction of individual responses to food
examining interindividual variability to nutrition inputs
show better prediction using algorithm, than simply knowing glycemic index of foods and meal composition
points to importance of understanding [physiological inputs and influences on metabolic responses
studies did not test whether giving individuals info helped them make diff dietary choices