BIN300 W10- application of GS
1/9
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
10 Terms
why do we expect so much from genomic selection
direct selection at DN level vs indirect for phenotype
paradigm shift: new way of thinking about selection, more accurate seleciton, people understand selection for DNA, not for phenotypes
accurate GEBVs of animals without records
can also have traditional EBV for animals without recs, but usually not very accurate, veyry important in some breeding schemes, e.g. selection of bulls indairy cattle breeding schemes
•EBV = estimate of breeding value; GEBV = genomic (selection) estimated of breeding value
•The ’accuracy of selection’ is the correlation between EBV and true breeding value (TBV).
• Selection for phenotypes is often poorly understood because people do not understand the relationship between phenotype and genotype. But in principle the models are not so different:
Phenotypic selection : y = genotype + environment
Genomic selection : GEBV = genotype + estimation_error
We expect that estimation_error is smaller than environmental effects.
• Traditional EBV of animals without records are based on the records of their parents, sibs or offspring.
•Since bulls obviously do not have milk production records, selection of bulls in dairy cattle breeding is traditionally based on progeny testing, i.e. bulls obtain about 100 daughters, and their selection is based upon the performance of the daughters.
Genetic improvement per year
genomic selection affects
accuracy of selection, but may be already high
genertation interval, genotyping for GS: immediately at birth, halving L → doubles delta G, L effect may be more important than r increase
•intensity of selection is related to how many you select out of how many candidates. 10 out 100 candidates gives better intensity than 50 out of 100 or 10 out of 50.
•σg is a measure of the size of the genetic differences (bigger genetic differences: more ΔG).
•L measures how fast you do the selection. More selection-rounds per decade : more ΔG.
•Reducing the generation interval is often what breeders makes enthusiastic about GS. But note: for some species the generation interval is already at its biological minimum eg. in pig breeding.

In the case of sequence data:
• In the case of sequence data: few QTL relative number of SNPs which implies that it is important to try to sift out which SNPs are the QTL. BayesB/C try to do this, GBLUP does not.
sequence dat ais more accurate than typical SNPchip
dairy cattle breeding
selection of young bulls (no proegny tests)
saves costs of progeny tests, reduce generation interval by factor 2, also uses GS on bull dams
double Delta G, saves money
•Progeny test in dairy breeding: let candidate bulls obtain ~100 daughters, record their performance, and select best bulls based on average performance of these test daughters. Very accurate test but takes >5 years before selected bulls get offspring.
•
•Much more genetic gain and cheaper => everybody very excited.
Fish breeding schemes
replace disease challenge tests on sibs by GS, esitmate marker effects from practical disease data, case-control deisng, GS in the nuclueus
no family housing needed, prblem, large number of selection candidates, so there needs to be a pre-selection for other traits
Disease challenge test in fish: split large fullsib family in two groups. Group1 becomes selection candidate and will be possibly selected. Group2 will be deliberately infected by a disease and their disease resistance is recorded. Because of the disease risk, all Group2 animals are destroyed. But Group1 animals may be selected based on the performance of the Group2 animals.
pig breeding schemes
GS of test-boars for maternal traits
estimate marker effects on sibs of test-boards
genotype test-boars and select for marker effects
carcass traits,
use crossbred practical performance info
record trait in practice on crossbreds, GS in the nucleus for crossbred perofrmance
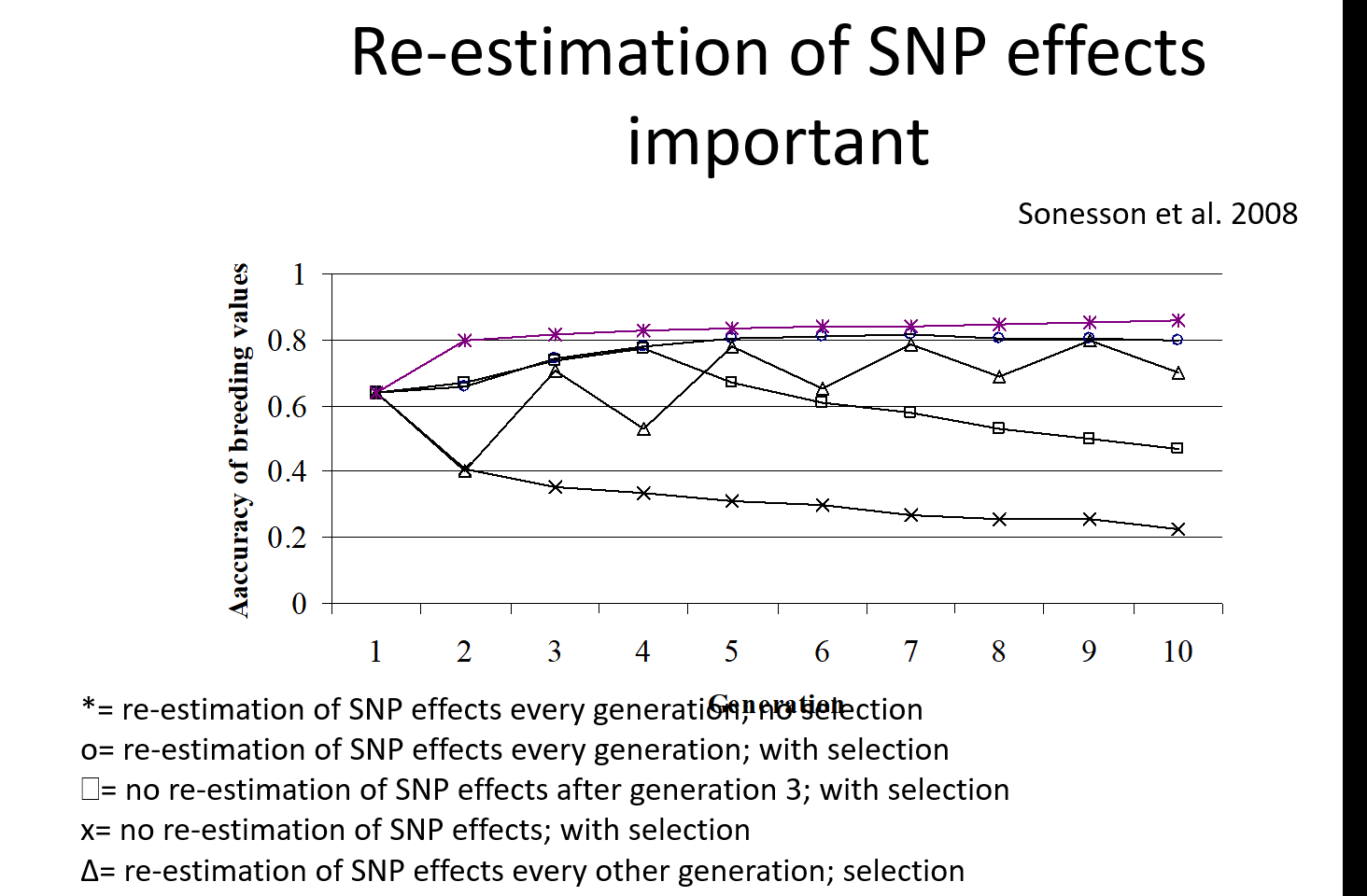
Can we estimate SNP effects once in a training opulation and selecti for these SNP effects for every afte?
no. genomic relationship between the training and canditate aniamls decrease over time as the candidates come from a younger and younger generation
Inbreeding and GS
GS improves prediction of within family effects, less weight on family effects, less co-selection of relatives → less inbreeding
other changes to the breeding structure can counteract the above effect
reduce generation interval, speeds up both annual inbreeding and genetic gain
selection at younger age: implies often less information on individual/within family effect•
• If family effects have a lot of weight in the EBV, then family-members have similar EBV and are all selected if their family effect is good. If there is little weight in family effects, the within family effects are important: if selected animal i comes from one family, then the next selected animal j can come from a completely different family, because within family effects are not related to the family where the animal comes from.

GS mainly important to predict EBV on non-phenoypted animals
can envisage different breeding structures, no recording of phenotype/pedigree o elite candidates, shortening of generation intervals, functional traits → more sustainable breeding
s
sequence data: 40% more accurate
accuracy persists for 10 generations