Transcription and mRNA processing
1/16
Earn XP
Description and Tags
know
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
Basics of transcription to produce mRNA
core promoter
enhancer
mediator
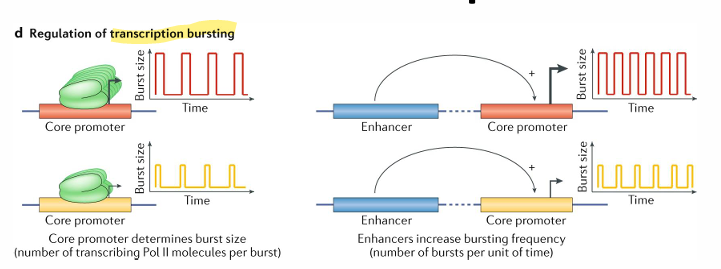
the bursting nature of transcription
RNA processing
5’ capping
Polyadenylation
RNA splicing to remove introns
Alternative splicing to modulate the functions of proteins
Distinct RNA Polymerases produce different classes of RNA
Pol II produces mRNAS, lncRNAs, and miRNAS
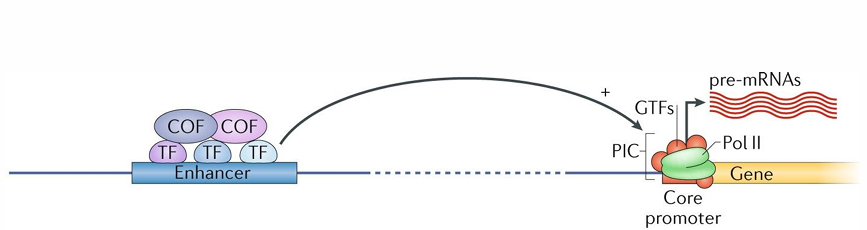
Simple model of transcriptional initiation
enhancer with transcription factors bound (which interact directly with DNA) and COF which activate transcription factors
GTFs: general transcription factors
PIC: pre-initiation complex (Pol II and GTFs) on gene
produces pre-mRNAs
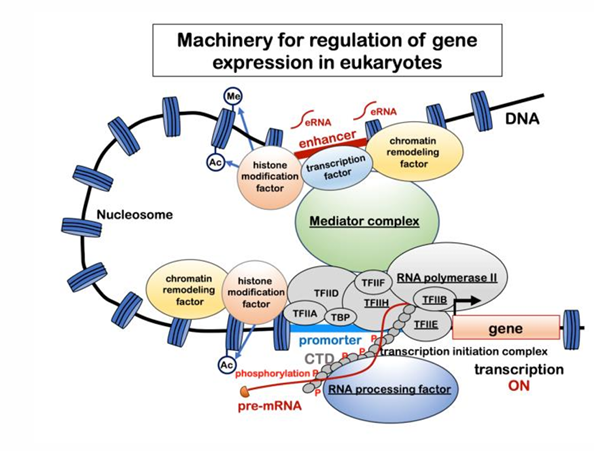
Histones, Looping, and Mediator complex for transcription
Histones: modification and remodeling (moving)
Looping: loops form between enhancers that can be 100,000s bp upstream or downstream of the transcriptional start and the PIC
Mediator complex: links the enhancer and polymerase complex
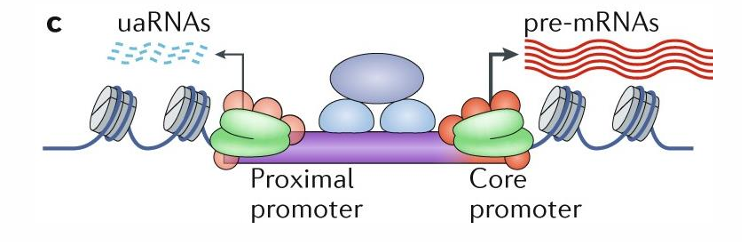
The ______ and __________ are the basic unit for initiation of transcription
proximal, core promoter
Cis elements within core promoters
NFR: nucleosome free region around the start sight (allows space for things to bind)
Elements are not in all promoters
Inr: 46% of promoters
TATA: 24% of human promoters
Sequence motifis within promoters and the proteins that bind them ***
***
Distinct types of promoters for classes of genes
a. Adult tissue-specific genes or terminally differentiated cell-specific genes
focused, sharp initiation
b. Housekeeping genes
expressed in all cells, dispersed, broad initiation
c. Developmental transcription factors
dispersed, broad initiation
two big (prominent) histones in promotors
Transcriptional initiation and elongation steps (4)
transcription factor binding to enhancer region
recruitment of co-activators which act on chromatin structure
PIC formation (the mediator helped facilitate the PIC assembly by looping the enhancer region to the promoter)
promoter escape
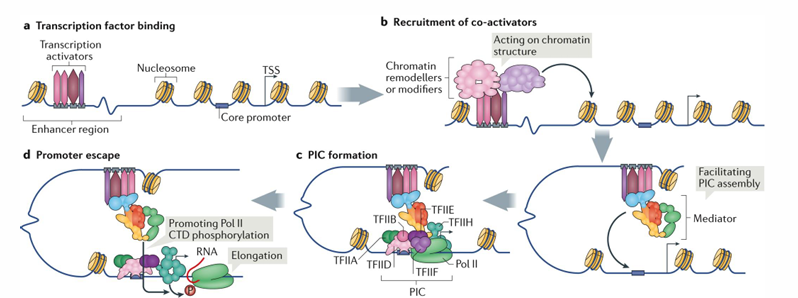
Overview of transcription and processing
there is promoter-proximal pausing that must be released
additional pausing at the first nucleosome that requires additional factors to proceed through
Transcription speeds
single cell techniques reveal that tehre are bursts of transcription that vary in the amount of RNA produced and the frequency of the burst
measurements of populations show smooth levels of transcriptions even though on the individual level this is not the case
it is thought that the core promoter regulates the burst size and the enhancer regulates burst frequency
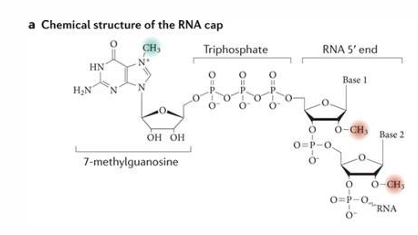
5’ Cap addition pros (5)
protects from degradation of 5’ → 3’ exonuclease
facilitates nuclear export
facilitates splicing
enhances translation
helps distinguish between self and non-self RNAs
Polyadenylation
PolyA stabilizes the mRNA
Alternative PolyA usage can change stability, localization, translation efficiency, and C-terminus of the protein
The spliceosome complex
splices RNA to remove introns
composed of RNA and proteins
individual components of RNA and protein are referred to as snRNP - small nuclear ribonucleoprotein
the RNA components are essential for the catalystic removal of the intron
The sequence of the cis elements are relatively degenerate
Alternative splicing
produces proteins with different modules to preform distinct functions
90% of human genes are alternatively spliced
alternative splicing is a mechanism for
genomic evolution
developmental processes
modulation of protein function
Regulators of alternative splicing
Cis elements in pre-mRNA regulating splicing
ESE – exonic splicing enhancer
ISE – intronic splicing enhancer
ESS – exonic splicing silencer
ISS – intronic splicing silencer
Two main families of alternative splicing regulators
SR – ser / arg rich proteins
hnRNP–heterogeneous nuclear ribonucleoproteins