Cellular and Molecular Biology - Term Test 1
1/123
Earn XP
Description and Tags
Lectures 1-9
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
124 Terms
bacteria
unicellular (mostly)
gram negative and positive
huge diversity of survival strategies
archaea
unicellular
many extremophiles
eukaryotes
unicellular and multicellular
protists, fungi, plant, animals
evolution of cellular life
cellular life evolved from a common ancestor
gene passed vertically from parent to child and horizontally within and between species
mitochondrion endosymbiosis gave rise to eukaryotes
primary endosymbiosis gave rise to photosynthetic eukaryotes
photosynthesis spread to other eukaryotes via secondary endosymbiosis
common characteristics of life
growth
metabolism
response to stimuli
movement reproduction
genome
the storage material containing the organisms genetic information
contains instructions for self-replication (genes)
all cellular life has double stranded DNA (dsDNA) genome
viruses can have a DNA or RNA genome, single-stranded or double-stranded (depends on the viral species)
all things necessary to survive (behaviours) come from genomes
central dogma
the underlying molecular mechanisms that controls life
controls all cellular processes such as growth, metabolism and response to stimuli
gene on the DNA genome is expressed through transcription, producing an RNA copy of the gene
what does transcription produce
depending on the gene, transcription produces mRNA or functional RNA
mRNA gets translated into proteins
functional RNAs do not get translated, they function as is
they take part in cellular processes as an RNA molecular
proteins and functional RNAs performs activities to regulate cellular processes
what are cells filled with
cells are a sack made of lipid membranes and are filled with aqueous solution containing various organic and inorganic molecules
macromolecules (nucleic acids, proteins, carbohydrates, lipids)
variety of other organic metabolite
inorganic ions
water
organelles (for eukaryotic cells)
prokaryotes
no membrane bound organelles
usually smaller than eukaryotes
mostly singled cellec
eukaryotes
has membrane bound organisms such as a nucleus
compartmentalizes biochemical reactions using organelles
usually larger than prokaryotes
single or multicellular
presence/absence of nucleus has a huge implication for transcription and translation
size of prokaryotic cells
1-2µm
size of eukaryotic cells
5-20 µm
size of mitochondria
1-2 µm
size of ribosome
20 nm
size of one atom
fraction of a nano-meter
the two types of microscopes
light and electron microscope
light microscope
uses photons to image structures
accessible and easy to use (at least of the simpler kinds)
can image live cells in real time
can image in colour
lower resolution
electron microscope
uses electrons to image structures
very high resolution
no real time or colour imaging
much more expensive and difficult to operate
archives a much higher resolution and can visualize organelles/large macromolecules
electron microscopes are harder to purchase and operate compared to a simple light microscopes
macromolecules
they are teh major structural, enzymatic and regulating components of cells
about 30% of bacterial mass/total volume are chemical compounds (dry meat)
about 80% of bacteria sage dry mass consists of macromolecules
types of macromolecules
sugars,
fatty acids
amino acids
nucleotides
phospholipids
two hydronic fatty acids, covalent bond to a hydrophilic head
phospholipid s non-covalently associated with themselves as via hydrophobic/hydrophilic.
phospholipid bilayer (heads facing up, tails facing down
sugars
polymerize into polysaccharides
polysaccharides used as components of cellular structures, energy storage, ect.
amino acids
polymerize into polypeptides (proteins)
proteins represent various cellular functions as much as metabolism, structure, regulation, signal transduction, etc
nucleotides
polymerize into nucleic acids
DNA is the storage material of genetic information
RNA can be na intermediate between DNA and protein (mRNA) or can have biological function by itself (function RNA)
sugars, animo acids and nucleotides always polymerize covalent bonds
nucleic acids are one of the four macromolecules
nucleic acids are long, linear polymer of nucleotides
RNA is a polymer of ribonucleotides
transient storage of genetic information (mRNA)
functional RNA
DNA is a polymer of deoxyribonucleotides
storage of genetic information in genomes
four types off deoxyribonucleotides used to build DNA
all four deoxyribonucleotides have similar composition with ‘bases’ that are slightly different from one another
adenine
thymine
guanine
cytosine
nucleotides structure
nitrogenous base + pentrose (five-carbon) sugar + one or more phosphate group
different kinds of nucleotides
nitrogenous bases
adenine, guanine, cytosine, thymine/uracil
pentrose sugar
ribose or deoxyribose
number of phosphates
1, 2, or 3 phosphates attached
purine (A and G)
purines have nine atoms constituting its two rings, numbered 1-9
nitrogen in the larger ring furthest away form the smaller ring is nitrogen 1
nitrogen in the smaller ring is connected to carbon 5 is nitrogen 7
nitrogen in the smaller ring without a double bond is nitrogen 9
the above is true at least for the two common purines used in the central dogma
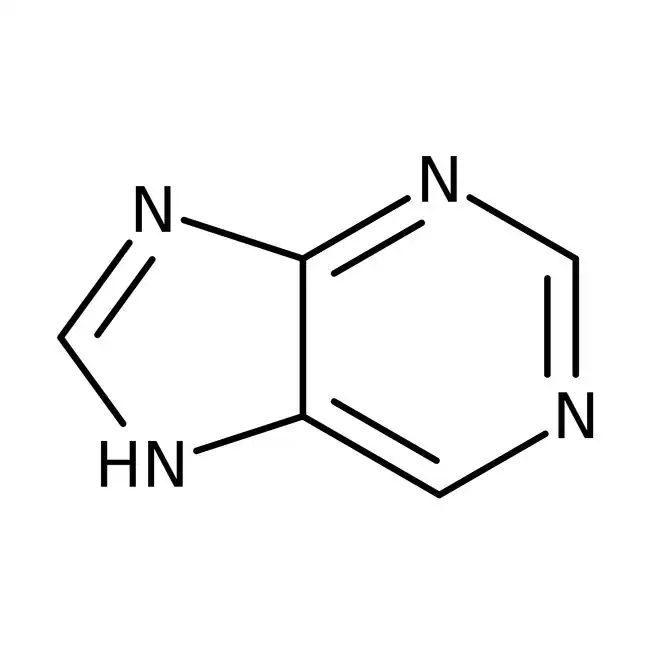
pyrimidines (T, C, and U)
have six atoms constituting its ring, number 1-6
two nitrogen’s at positions 1 and 3
carbon 2 is always connected to an oxygen with a double bond
carbon 4 is always connected to an extra atom outside the ring
the above is true at least for three common pyrimidines used in the central dogma
two types of pentose sugars (both a ribose)
carbons are numbered 1’ to 5’
ribose
has OH groups attached to its 2’ and 3’ carbons
found in RNA
deoxyribose
has an OH group attached to its 3’ carbon
missing the OH group at its 2’ carbon, compared to ribose
found in DNA
up to three phosphates are added
phosphate groups are added to the 5’ carbons found of the pentose sugar
the phosphates are named alpha (a) and beta (b) and gamma (v)
alpha is the closest to the 5’ carbons
gamma is the furthest away
a lot of energy is stored inside the phosphoanhydride bonds between gamma-beta and beta-alpha phosphates
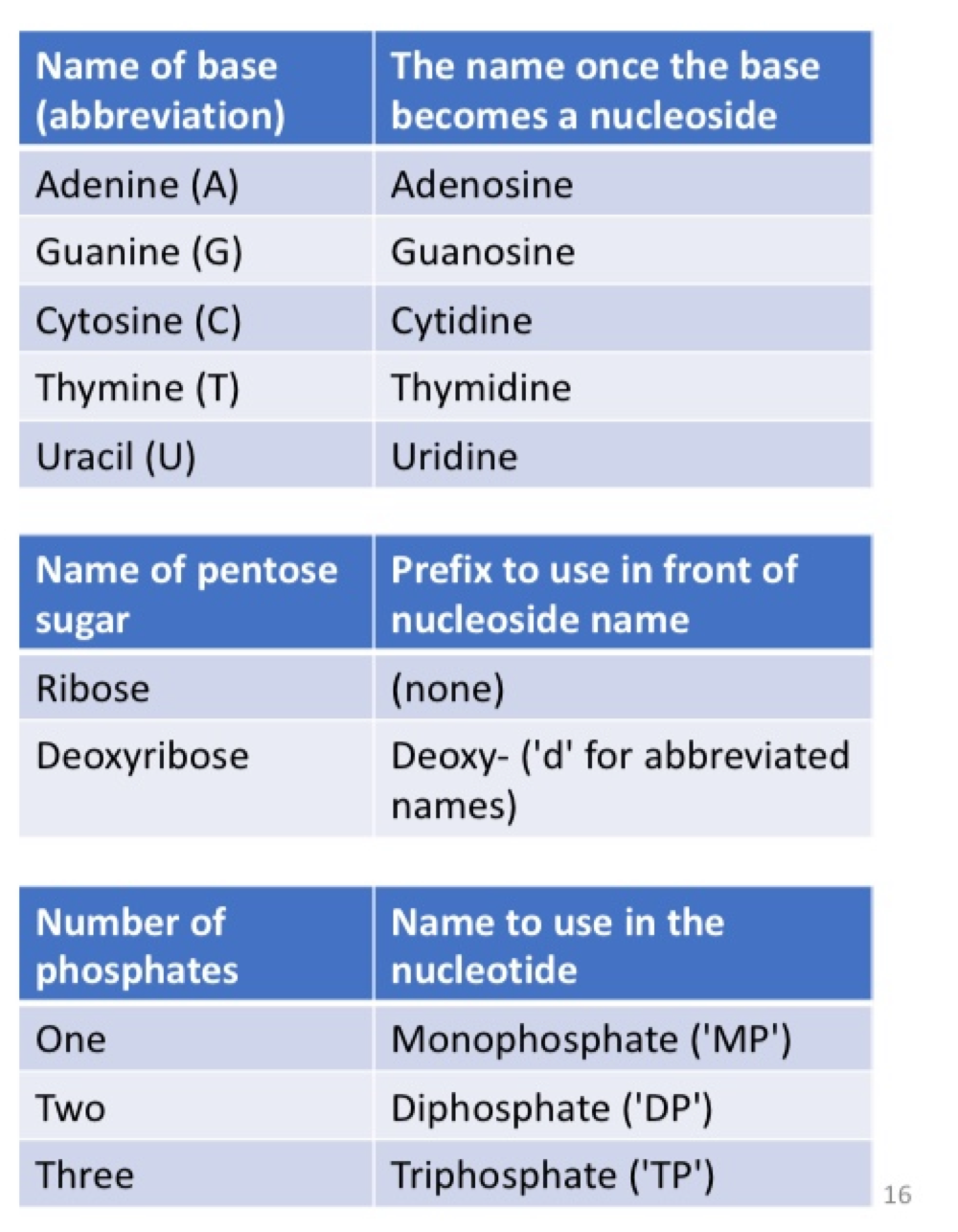
nucleoslide
nitrogenous base + pentose sugar (without any phosphate)
nucleotide
nitrogenous base + pentose sugar + at least one phosphate
nucleotide nomenclature
we follow the IUPAC code to name and abbreviate nucleotides systematically
the letter ‘N’ is used to represent ‘aNy’ nitrogenous base in DNA
in a DNA sequence, writing ‘N’ means that the position can be any one of A, G, C, or T
abbreviation for other common combinations of nucleotides:
‘R’ = A or G (puRines)
‘Y’ = T or C (pYrimadines)
nucleotide triphosphates
the incoming nucleotide must always be a triphosphate
the 3’ OH attaching to the alpha-carbon of incoming nucleotide trisphosphate breaks the high energy phosphoanhydride bond between beta-alpha phosphate
breaking bond between beta-alpha phosphates release
energy
pyrophosphate
get broken to release more energy
this energy is then used to catalyze the formation of the 3’ - 5’ phosphodiester bond
nucleotides are energy currencies
phosphoanhydride bonds in nucleotides can be broken
nucleotide polymerization is a special case
the reagent of reaction (NTP or dNTP) carries the energy to polymerize itself
therefore nucleic acid synthesis must use triphosphate nucleotides as reagents
mono- and di- phosphate nucleotides cannot be used during nucleotide polymerization since they don’t carry enough energy
nucleotide has many functions in the cell
being the building block of DNA and RNA is a major function of nucleotides
storage of genetic information, various activities as functional RNA
other critical function is energy currency
enzyme currency
breakage of high energy phosphoanhydride bonds are coupled to energetically unfavourable reactions
ATP and GTP are major nucleotides used as energy currencies
ATP and GTP are generated using energy released by digesting food
coenzymes for various metabolic pathways
coenzyme A
adenosine derivative with a 5’ di-phosphate and a 3’ monophosphate
5’ di-phosphate connected to more structures
coenzyme A is used to facilitate many metabolic pathways
fatty acid metabolism
gets added to private to Acetyl-Coft, which is the starting point into the citric acid cycle (TCA cycle)
electron carrie for metabolism
NAD+ is dinucleotide
first nucleotide has an unconvential base, nucliotinamide (this is NOT a pyrimidines)
second nucleotide is an AMP
two nucleotides attached directly via alpha phosphates
NAD+ receives electrons released during glycolysis and citric acid cycle
this converts to NAD+ and NADH
electrons donated to the electron transport chain to generate ATP
cAMP is made from ATP
bond between alpha-beta phosphates broken
the open end of alpha phosphate gets connected to the 3’ carbon of its own ribose sugar, making a cyclic structure
formation of double stranded DNA (dsDNA)
nucleic acids are synthesized via polymerization of nucleotides
polymerization of dNTPs generate single-stranded DNA (ssDNA)
synthesis in the 5’ to 3’ direction
long, unbranched chain of nucleotides
in our cells, DNA rarely exists as ssDNA
ssDNA attached to form a second, complementary ssDNA to form double stranded DNA
dsDNA bonds
covalent bonds hold nucleotides in ssDNA (3’ - 5’ phosphodiester bonds)
hydrogen bonds hold two ssDNA together in dsDNA
no covalent bonds occur between two strands of ssDNA
the hydrogen bonds of dsDNA occur between nitrogenous bases of ssDNA
nitrogenous bases are held inside the middle of dsDNA
sugar phosphate backbone occur on the outside
two ssDNA in dsDNA associate in an anti parallel orientation, where their 5’ - 3’ directionally is reversed
base pairing
nitrogenous base always pair with a specific partner
cytosine pairs with guaine via three hydrogen bonds occur
thymine pairs with adenine via two hydrogen bonds
these are called base pairs (bp)
base pairs are written as G.C and A.T
full chemical structure of dsDNA
these G.C and A.T base pairs are part of a longer stretch of dsDNA
5’ and 3’ ends of this dsDNA are connected to more nucleotides (dashed lines)
numbers are shown for important atoms in red
distance between two C1’ carbons in a base pairs is about 1.1 nm
in the true 3D structure, nitrogenous bases and their sugars are held perpendicular to each other
physiological implications of base pairing
the G.C has one more hydrogen bond compared to the A.T pair
it takes more energy to break apart a G.C pair
G.C pair holds the two strand of dsDNA more strongly compared to the A.T pair
for example, dsDNA with higher GC% is harder to break into ssDNA using heat (desaturation)
GC% of the genome is species dependent and is highly variable
prokaryotes have gemones with GC% of 8-75%
organisms living in a higher temperature tends to have genomes with a higher GC% presumably to resist denaturation by heat
calculating GC% and AT% of dsDNA
since dsDNA has complementary base pairing
for every adenine, there is a corresponding thymine
for every guaine, there is a corresponding cytosine
therefore A% = T% and G% = C%
base pairing is the basis of DNA…
during DNA replication, two strands of dsDNA are broken apart and used as a template to help synthesize new strands of DNA
base pairing is also the basis for transcription (generates ssRNA using ssDNA as a template)
writing DNA/RNA sequences
nucleotide sequences are ALWAYS written in the 5’ to 3’ direction
DNA/RNA sequences written without a label are assumed to be written in this direction
if you must write them in a 3’ to 5’ direction you must make this very clear
for dsDNA, chose on of the ssDNA sequences to represent the whole molecules
it is not necessary to indicate both sequences as they are complementary to each other
show the sequence of the more important ssDNA (for example, the coding strand of a gene as opposed to the non-coding strand)
discovery of the dsDNA structure
phoebus aaron theodore levine
erwin chargaff
x-ray crystallography
under supervision of rosalind franklin, raymond gosling took x-ray diffraction images of crystallized dsDNA (early 1950s)
the 51st image (photo 51) was shown to Maurice Wilkins, who then showed it to James Watson
James Watson and Francis Crick solved the structure of dsDNA using photo 51 and published in 1953
phoebus aaron theodore levene
multiple discoveries between 1910s - late 1920s
showed that DNA contains equal proportion of nitrogenous bases, deoxyriboses, and phosphates
hypothesized that a nitrogenous base, deoxyribose and phosphate combine to make the building block of DNA, and named this unit ‘nucleotide’
Erwin Chargaff
discovered the Chargaff’s rule, late 1940s
in dsDNA, around of guanine = amount of cytosine, and amount of adenine = amount of thymine
X-ray crystallography
method to analyze the atomic structure of a crystal
can be used to analyze structures of macromolecules like proteins and nucleonics acids
purity and crystallize molecules
shoot x-ray through the crystal
the crystal diffracts x-ray and the pattern is recorded on a film/screen
diffraction pattern represents the structure of the crystal = atomic structure of molecule in the crystal
physical dimensions of dsDNA
an antiparallel, double stranded helix with a right handed twist
dimeter = 2nm
length occupied by one base pair = 0.34 nm
length of one turn = 3.4 nm
therefore = 10 base pairs per turn
B-DNA is physiologically relevant form of dsDNA
dsDNA can fold in different shapes under different conditions
B-DNA is the physically relevant, hydrated from
right-handed helix
occurs in physiological conditions (chemical conditions inside our cells)
A-DNA is the ‘dried’ form
right handed helix
occurs at <75% humidity
left handed Z-DNA may occur in cells, but function is not well understood
dsDNA structure
base pairs in the middle
sugar phosphate backbone on the outside
base pairs in the middle are not completely hidden by the backbone
openings between the backbone exposes some parts of base pairs
openings between sugar-phosphate backbones are not evenly spaced
nitrogenous base pairs are laid flat when you look down at dsDNA along its length
two deoxyribose stick out from the base pairs in a non 180º angle
one side of the base pairs has a wider opening (257º)
opposite side has a narrower opening (103º)
these openings become the major and minor grooves of dsDNA, when these base pairs stack onto each other (while rotating 36º every bp since 10 bp = 1 turn)
major groove
larger gap between the backbone exposes
expose the bases more due to its wider opening
minor groove
smaller gap between the backbone and
major and minor grooves
major and minor grooves always occur in a pair, and on opposite sides of dsDNA
major and minor grooves expose some atoms of nitrogenous bases to the environment
‘environment’ = nucleus, cytoplasm, etc.
major and minor grooves allow other macromolecules (like proteins) to directly touch the nitrogenous bases
every nitrogenous base has a unique shape
proteins can tough and READ the sequences of dsDNA without opening the dsDNA
enables efficient protein DNA interaction when DNA binding proteins are searching for specific DNA sequences to bind to
it takes time and energy to open up dsDNA to completely expose the bases
discovery of the basic feature of RNA
in the 1953 paper, Watson and Crick described dsDNA structure without surprisingly occuract
they even correctly, hypothesized the machinist of DNA replication, just based on the dsDNA structure
however, they made some claims that turned out to be incorrect
watson and crickets hypothesized that RNA can not fold into a structure similar to B-DNA
in reality, dsDNA looks very similar to B-DNA
basic features of RNA
RNA folding into double stranded structures are the basis of many molecular mechanisms
chemical composition and polymerization mechanism or RNA and DNA are nearly identical
structural difference between RNA and DNA
ribose is used as the pentose sugar in RNA
uracil is used in the place of thymine in RNA
RNA usually exists as ssRNA in the cell
ssRNA can fold onto itself by complementary bases-pairing
ssRNA folds by base pairing
ssRNA is a long, flexible string
ssRNA can base pair onto itself using complementary base pairing
unique shape of folded ssRNA gives it biological function (RNA)
in HIV< a portion of its mRNA folds into a unique shape (RRE) to bind proteins that guide the mRNA for translation and packing
RNA Function
not for permanent storage of genetic information in cellular organisms
some viruses such as HIV, coronavirus and Ebolavirus have RNA genomes
multiple physiological functions in the form of mRNA, tRNA, rRNA, snRNA, etc
functional RNAs do not get translated into proteins
fold into unique 3D shapes to perform their functions
common characteristics of life
polypeptides are long polymer of animo acids
polypeptides fold into proteins
proteins participate in diverse cell functions
enzymes
structural components
signaling
receptor
motor proteins
transport systems
gene regulation
immune systems and antibodies
amino acids are the building blocks of polypeptides
20 types of amino acids are used to build polypeptides
all 20 amino acids have similar compositions with different side chains attached to the amino acid backbone
the 20 bases can be connected in any number/order
action acid sequence of the polypeptide determines how it folds
amino acid structure
amino acids has a central carbon (alpha carbon, Ca) connected to 4 ‘groups’
amino group
carboxyl group
side-chains group (R-group)
hydrogen
R-group is the variable component between different amino acids
all other components are the same (except for proline)
at physiological pH, amino group and carboxyl group becomes positively and negatively charged, respectively
side chains determine properties of amino acids
r group varies in
size and shape
hydrophobicity/polarity
charge
chemical reactivity
amino acids used by cells to build proteins `
10 amino acids are non polar
10 amino acids are polar
5 of these are not charged
2 of these carry a negative charge
3 of these carry a positive charge
amino acids with non polar side chains
10 in total
R group is usually a hydrocarbon (carbons and hydrogens)
pure hydrocarbons are not polar
amino acids with polar, uncharged side chains
5 in total
R group contains an amide or hydroxyl (OH) group
amines and hydroxyls have electrons unevenly distributed within their structure
this makes one part of the structure more positive, and other parts more negative (polar)
2 amino acids ahve polar, acidic side chains attached physiological pH
donates a hydrogen and becomes negatively charged
in both cases, the acidic group is a carboxyl group
3 amino acids have polar, basic side chains at physiological pH donates
accepts a hydrogen and becomes positively charged
amino, guanidum or an imidazole group
amino acids a re categorized mainly by their polarity
polarity determines how each amino acid side chains interaction with water
water is a polar molecule
have uneven distribution of electrons
one side is slightly negative, on their side is slightly positive
polar side chains like to be beside water since they also have an uneven distribution of electrons, creating a positive/negative surface that attracts water
non polar side chains dislikes being beside water because these side chains has an even distribution of electrons
there is a varying degree of hydrophobicity even between amino acids in the same category
some amino acids have hydrophobicity that are in-between polar and non polar
amino acids with unique properties: one that influence folding very strongly
how a polypeptide fold into a protein is determined by its sequence of amino acids residues
polarity of each amino acid residues have huge influence in this process
certain amino acids have additional features that strongly impacts protein folding
glycine: smallest amino acid, makes the polypeptide backbone very flexible
proline: has a side chains that connects to its nitrogen, makes the polypeptide backbone very inflexible
cysteine: can form di sulphide bonds with other cysteine residues to covalent link tow polypeptide backbones
amino acids with unique properties: ones with aromatic rings
aromatic rings are larger and bulkier compared to other side chains
takes up more space when proteins fold
tyrosine, phenylalanine and tryptophan has side chains that absorb ultraviolet at 270 - 280 nm
polypeptides that contain these amino acid residues can be analyzed by UV-spectrophotometer
histidine has an imidasole ring that has a pKa of ~6.0
therefore, only some histidine residues are positively charged at physiological pH
other histidine residues are uncharged
small changes in pH dramatically affects this behaviour of histidine
the ‘flexibility’ of histidine side chains are utilized by enzymes to handle protons on/off the other
amino acids with unique properties: ones with a hydroxyl group
serine, threonine, and tyrosine has a hydroxyl groups exposed at the end of their side chains
exposed hydroxyl groups are great targets for phosphorylation
kinase: any enzyme that adds phosphate group to target
phosphate: any enzyme that removes phosphate group from target (dephosphorylation)
phosphorylation is used to regulate protein function after it has been made
for example, some proteins are only active when they are phosphorylation and vice versa
cells phosphorylation/dephosphorylate their proteins to switch them on and off in response to their physiological needs
amino acids polymerize into polypeptides
the end of a polypeptide has a free carboxyl groups exposed
this is called the carboxyl terminus (C-terminus) of the polypeptide
next amino acids is placed beside the polypeptide
amino group of the amino acid is facing the carboxyl group at the C-terminus of polypeptide
condensation reaction joins the carboxyl carbon of the polypeptide to the nitrogen of the next amino acid
this newly formed bond is called the peptide bond
polypeptide has a N-C-C backbone
individual amino acids (before polymerizing have nitrogen, alpha carbon, and carboxyl) upon polymerization, these three atoms form a long chain with a repeating ‘NCC patter’
this is the peptide backbone
N-terminus (amino-terminus)
the front of the peptide backbone with a free amino group
C-terminus (carboxyl-terminus)
the end of the peptide backbone with a free carboxyl group
amino acid vs amino acid residue
the chemical structures of amino acids change when they polymerize
inside the polypeptide they are no longer ‘amino acids’ are still recognizable inside the polypeptide
individual amino acids are still recognizable inside the polypeptide
these ‘remainders’ of amino acids are called amino acids residues
the polypeptide in this figure is made out of four amino acid residues
polypeptide vs proteins
polypeptide is any chain of amino acids polymerized via peptide bond
protein is defined more vaugely
protein is a longer polypeptide that folds into a unique 3D shape and obtains a biological function
proteins are usually over 100s of amino acid residues long
peptides fold into proteins
conformation: a proteins final folded structure
polypeptide must fold in a shape that satisfies the needs of its amino acid residues
polar sides chains facing water
polypeptide folding is determined by its sequence of amino acid residues
protein structure is dynamic
a protein may have multiple stable conformations
switches from one conformation to another while it performs its biological activity
polypeptide folding in aqueous environments
proteins that function in aqueous environment are surrounded by water
inside the cytoplasm, nucleolus, ER, golgi, mitochondria, etc
these proteins usually fold into a sphere
place hydrophilic amino acid residues on the surface of the protein, exposing them to water
police hydrophobic amino acid residues inside the protein, away from water
becomes the hydrophobic core
polypeptide folding is supported by three non-covalent interactions
van der waals attractions
very weak interaction when atoms are placed closer to another
hydrogen bonds
bonding between two polar molecules
electrostatic interactions - bonding between two charged molecules
these interaction are weaker than covalent bonds, but they occur in large number and accumulate into a strong force
unique amino acids the influence protein folding: cystine
no covalent bonds contribute to protein folding except for one example
a pair of cystine side chains can form coavalent disulfied bonds to connect the polypeptide backbone
holds protein structure very strongly
antibodies are produced by our immune system to bind and remove potentially harmful materials (antigens) in our body
two antigen binding sites form at the ends of the antibody
one antibody is made of four individual polypeptides
two units of ‘heavy chain’
two units of ‘light’ chain’
multiple disulphide bonds contribute to this structure
disulfide bonds can occur
within the same polypeptide backbone
between two different backbones
flexibility of polypeptide backbone
the polypeptide backbone is flexible
allows it to hold into many shapes
not all parts of the polypeptide backbone are equally flexible
the bonds before and after the central carbon are flexible
N-Ca bone and the Ca-carboxyl
peptide bond is inflexible
protein folding is limited by these properties
unique amino acids for folding: glycine
glycine is the smallest amino acid
side chains is a single hydrogen
glycine increases the flexibility of surrounding area since it provides more ‘open’ space for other atoms to occupy
mutations that replaces amino acids into a glycine may destabilize protein structures because it introduces flexibility
unique amino acids for folding: proline
proline has its side chains covalently attached to its backbone nitrogen
its technically not an ‘amino’ acid since it does not have an amino group
proline is an amino acid
prolines N-Ca bond is completely inflexible as its rotation is locked by the side chain
mutations that replaces amino acids into a proline may disrupt protein structure by removing the flexibility that was necessary to accomodate the original fold
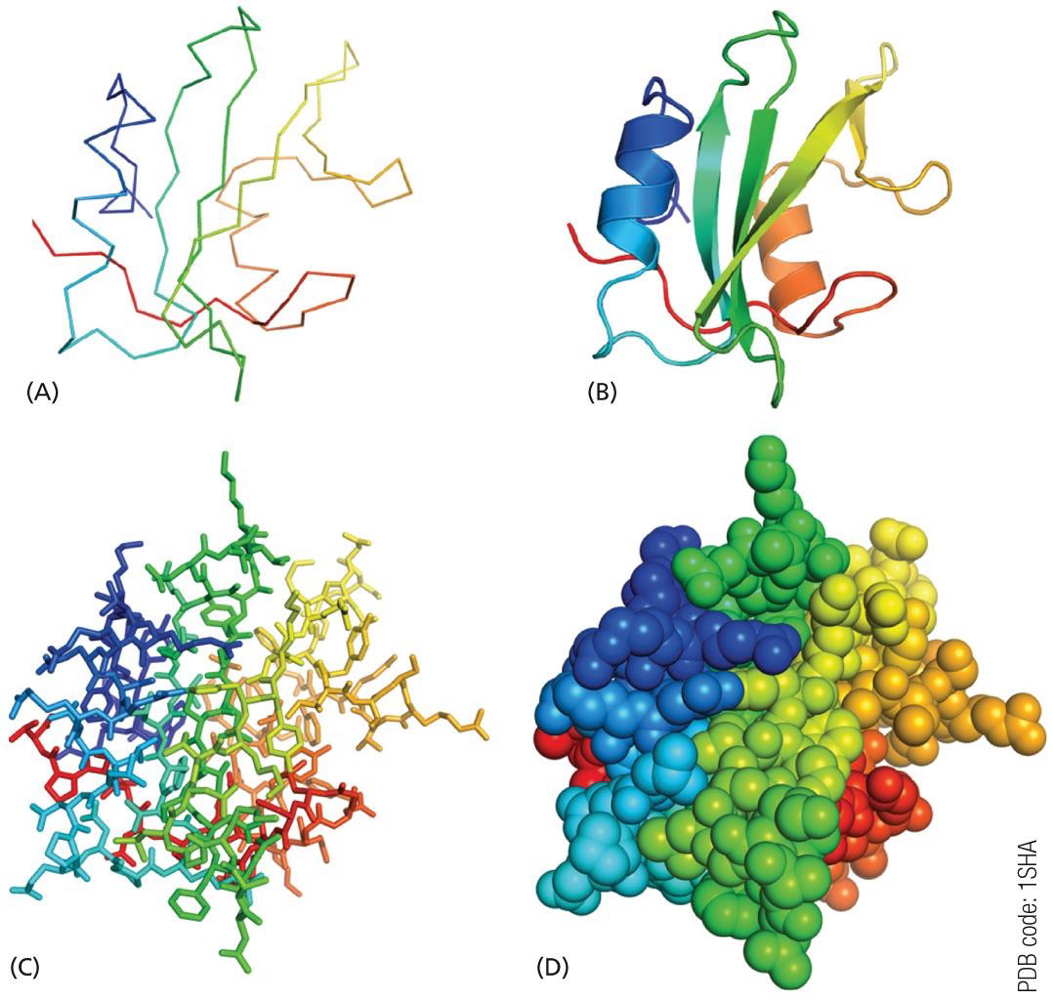
protein structures can be represented in many ways
use appropriate method to best represent what you want to show
A. peptide backbone represented as lines
B. peptide backbone represented as ribbons to show secondary structures
α helix: curled ribbons
β sheet: ribbon with arrows
C. all atoms shown as sticks to display the side chains
clutters the image but can represent molecular interactions using a specific side chains groups
D. all atoms shown using a space filling model
represents the surface shape of the protein
protein data bank
scientists have been analyzing atomic structures of biological molecules for about a century
x-ray crystallography
nuclear magnetic resonance (NMR)
cryogenic EM
structural data deposited in the protein data bank
open access
structure catalogued with a four letter code
over 200,000 structures are currently deposited
protein structure is described by four level of ‘complexity’
primary structure (1º)
linear amino acid sequence of the protein
secondary structure (2º)
short regions of the peptide backbone fold into individual 3D structures that compose the tertiary structure
tertiary structure (3º)
the fully-folded, 3D structures of the protein
quaternary structure (4º)
in many cases, multiple individual proteins combine into a larger, mega structure
primary structure (1º)
primary structure is the raw sequences of amino acid residue of the protein secondary structure
also called ‘primary sequence’ ‘amino acid sequence’
written from N-terminus to C-terminus
primary structure determines how the polypeptide folds
sometimes, a change in single amino acid change have a large effect on protein function
multiple sequence alignment (MSA)
compare primary sequences of different proteins
mutant protein vs. wild type
homologous proteins from different organisms
can also compare nucleotide sequences
this example compares two mutant proteins against the wild type
mut_1 has the 4th valine substituted with leucine (non polar to non polar)
mut_2 has the 4th valine substituted with glutamic acid
we can hypothesize the mut_2 is affect more because its mutation substitutes valine into something completely
nomenclature of MSAs and Mutation
in MSA an asterisk (*) indicates that all proteins have the same amino acid residues at that position
writing mutation
for example, mutation is mut_1 is written V4L because it has valine at position 4 changed to leucine
mut_2 has a V4E mutation
mutations in DNA can be written the same way
secondary structures (2º)
proteins fold into their final shape (conformation) in a cell
this is their tertiary structure
tertiary structure contains multiple substructures that have distinct shapes
these substructures are called secondary structures
proteins do not fold first into secondary structures followed by tertiary
they fold directly from primary structure into tertiary structure
secondary structures do not occur independently
secondary structures are the ‘building blocks’ that combine into a tertiary structure
dissecting tertiary structures into secondary structures enable us to understand proteins better
folding of the peptide backbone
secondary structure is the folding of the peptide backbone
primary sequence (= sequence of side chains) determines what type of secondary structure that backbone becomes
however, side chinas do not directly stabilize the secondary structure by forming bonds, etc
hydrogen bonds between backbone atoms hold the secondary structure together
α-helix and β-sheet are the two major secondary structures
310 helix, π-helix, various turns and loops also exist