MCB Lecture 10 - Population Genetics and Evolution
1/62
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
63 Terms
Are allele frequencies constant?
NO.
Populations have different frequencies of alleles
In widely dispersed populations, allele frequency varies within sub populations
Ex: Human Brown vs. Blue Eye Color
What is an example of allele frequency inconsistency
Ex: Human Brown vs. Blue Eye Color
Why do allele frequencies change?
Natural populations rarely meet Hardy-Weinburg assumptions
NO POPULATION IS INFINITELY LARGE
New mutations at each locus arise occasionally
Migrations of small groups of individuals does occur
Mating isn’t random
Genotype-specific differences in fitness
What is the Hardy-Weinberg equation useful for?
Estimating population changes through a few generations
(Not as useful for predicting long term changes but does provide foundation for modeling)
What do Monte Carlo Simulations do?
Perform random sampling of inputs to produce probability distribution
Useful for modeling systems where inputs have high uncertainty or the mathematics is intractable/too complex
Named after Monte Carlo Casino
What is Monte Carlo Simulations specifically used for in Population Genetics
Used to model changes in allele frequencies
Starting population has a defined number of individuals that are homozygous/heterozygous
Experiment runs for specified number of generations
Multiple, independent experiments performed
At each generation in the simulation (4 things - Monte Carlo)
Random mating pairs chosen
Genotypes of offspring based on probabilities
Continues until total offspring number = parental population size
Parental generation is discarded, offspring are parents of next generation
Monte Carlo - What does each simulation represent?
A possible pathway of genetic drift
What is Genetic Drift?
Change in allele frequencies as a consequence of randomness in inheritance due to sampling error from one generation to the next
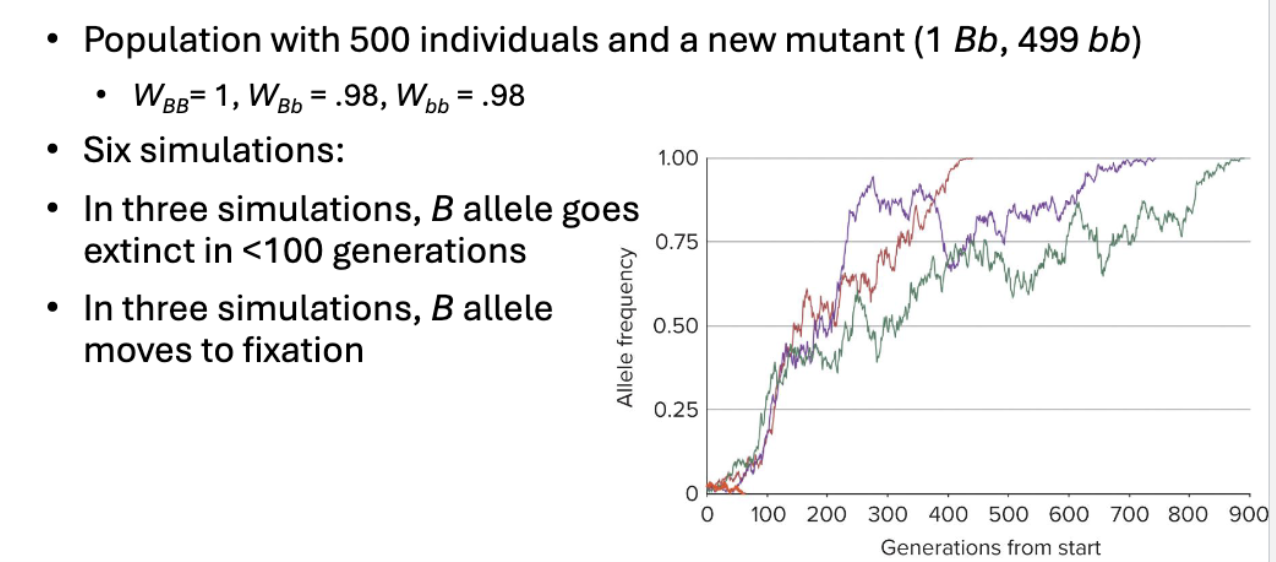
Six Monte Carlo simulations were run with 2 initial populations of heterozygous individuals, what happened?
NO SELECTION
Small population with 10 individuals had more genetic drift than larger population with 500 individuals
What is Fixation?
Fixation: When only one allele in a population has survived and all individuals are homozygous for that allele
Fixed because no further changes can occur (in absence of migration of mutation)
How big are the changes in allele frequencies each generation?
Relatively small
Over many generations can accumulate (once tiled, easier)
What is the Founder effect?
A few individuals separate from a larger populations and establish a new population
What are population bottlenecks?
A large proportion of individuals die, resulting in a smaller population
What are population bottlenecks?
A large proportion of individuals die, resulting in smaller population
What happens in both founder effects and bottlenecks?
A small subset is taken from a large population and then grows
2 places to introduce allele change:
Random subset may not be representative
Small population causes rapid genetic drift
What are mutations?
Variant DNA sequence in individual genome that wasn’t present in either parent
Deleterious: Reduce fitness
Beneficial: Increase fitness
Neutral: No benefit/harm
What is molecular clock?
Mutations accumulate in populations at fairly constant rate over time
DNA differences between organisms can be used to estimate how long ago they shared common ancestor
How does mutating allele affect allele frequency?
VERY DIRECTLY
EXTREME EX: 100% AA individuals in population, single mutation makes it 99.999% AA, 0.01% Aa
ANOTHER EX: 25% A1A1, 50% A1A2, 25% A2A2, mutation adds a new allele A3, changing all 3 frequencies
When generally isolated populations exchange individuals…
Allele frequencies and ratios can change!
(This is most noticeable when a new allele is added)
The amount of change in allelic frequency due to migration between populations depends on:
Difference in allelic frequency
Extent of migration
What are examples of nonrandom mating?
Positive assortative mating
a tendency of phenotypically like individuals to mate
Negative assortative mating
a tendency of phenotypically unlike individuals to mate
Inbreeding vs. Outcrossing
Crossing btw. closely related vs. unrelated individuals
(Based on genotype, not phenotype)
Inbreeding coefficient
Probability that 2 alleles at any locus are identical by descent of a common ancestor of 2 parents
Inbreeding depression
Reduced fitness due to loss of genetic diversity
Ex: Corn, loss fitness and smaller corns, fewer seeds, etc)
What is positive assortative mating?
Positive assortative mating
a tendency of phenotypically like individuals to mate
What is Negative assortative mating?
Negative assortative mating
a tendency of phenotypically unlike individuals to mate
What is inbreeding v. outcrossing?
Inbreeding vs. Outcrossing
Crossing btw. closely related vs. unrelated individuals
(Based on genotype, not phenotype)
What is inbreeding coefficient?
Inbreeding coefficient
Probability that 2 alleles at any locus are identical by descent of a common ancestor of 2 parents
What is inbreeding depression?
Inbreeding depression
Reduced fitness due to loss of genetic diversity
What are the effects of inbreeding on homozygosity?
Starting at 50% homozygotes in population rapidly reduces heterozygosity
Basic math: prob of 2 siblings sharing given allele from 1 parent is ½ so change of passing down is ¼
Cousins: 1/16, slower but still non-random
Deviation from HWE that AA and aa frequency are higher than expected from p and q
Elaborate on inbreeding/homozygosity
When progeny, the change of them being homozygous is 0.36 for p and 0.16 for q and the heterozygote is remainder
When inbreeding, odds become more skewed, if homozygote and relative, the odds that the relative homozygous for same allele higher
it’s also more likely to be a heterozygous - higher than expected
inbreeding, closely related they are, the more likely to be homozygotes in their progeny than population at odds
What is fitness?
An individual’s relative ability to survive and transmit its genes to the next generation
Statistical measure of all individuals with that genotype in population not a single individual
Two basic components: viability and reproductive success
Represented as probability from 0-1 where 0 is no change of offspring and 1 is guaranteed offspring
What is Natural Selection?
Process that increases fitness over generations
Individuals whose fitness is higher are parents of larger portion of next generation
Occurs in all natural populations
What is an example of Natural Selection?
Pocket mice in New Mexico live in rocky areas
Mice living in sandy rock areas tend to have sandy colored fur
Mice living in dark, lava rock areas tend to have darker fur
Likely due to increased predation of individuals that don’t blend

What is Sexual Selection?
Evolution is driven by mate choice or competition for mates
Sexual selection rapidly drives shifts in allele frequency by violating 2 HWE assumptions:
Non-random mating
Impact on fitness
Directly impacts the reproductive success, sometimes at expensive of viability

How does fitness and hardy weinburg intertwine?
Once fitness is involved, HWE no longer possible
However, Hardy-Weinburg formula can still make predictions
Ex Population with 2 alleles (A and a)
Relative fitness (W) of each genotype
Relative frequencies of each at adulthood
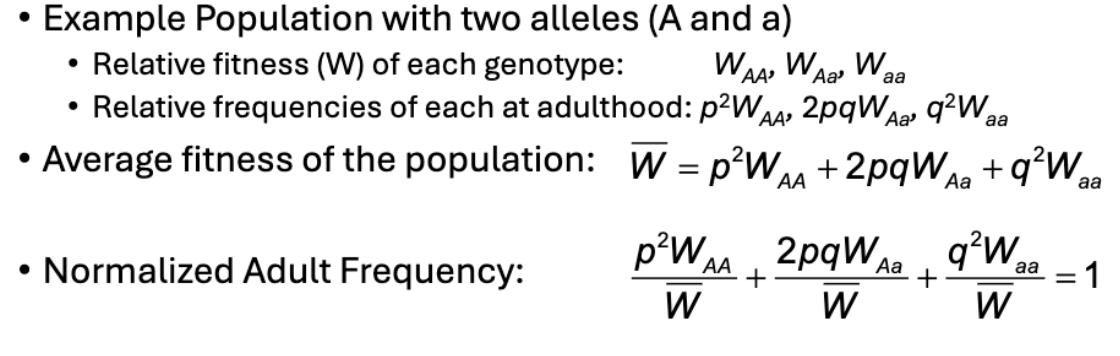
What are relative fitness used to determine?
The effective adult population size/frequency
If you have 100 AA individuals with 0.75 fitness, then only 75 will reproduce
After fitness is applied, genotype frequencies no longer add up to 1
What is the Normalization Factor?
The effective total adult population size/frequency and is used to make frequencies add to 1 again
What does p’ and q’ represent?
Allele frequencies after 1 generation of selection
What is the formula for change in allele frequencies
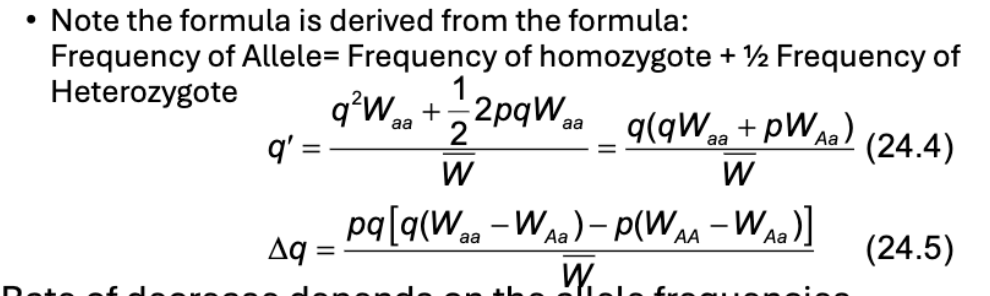
What does the rate of decrease depend on?
Allele frequencies
As q approaches 0, what happens?
Rate of decrease gets slower
Think of an example with recessive lethal and low q.
As 1 decreases, the frequency of homozygotes born decreases
So as long as there is no selective disadvantage for heterozygote, those alleles are “safe” from selection
With low q, heterozygotes are more likely to mate with p homozygotes
In a population with 500 individuals and 1 new mutation…
What are some human examples?
H. sapiens migrated out of Africa 60,000-80,000 years ago
Exposure to UV rays decreases with increasing distance from equator
Affects Vitamin D production and skin cancer incidence
Close to equator, dark skin protects against skin cancer
Farther from equator, lighter skin allows more UV for sufficient vitamin D production
Skin pigmentation is a complex quantitative trait determined by alleles at many genes
Alleles of several genes show strong associations with different populations around the world
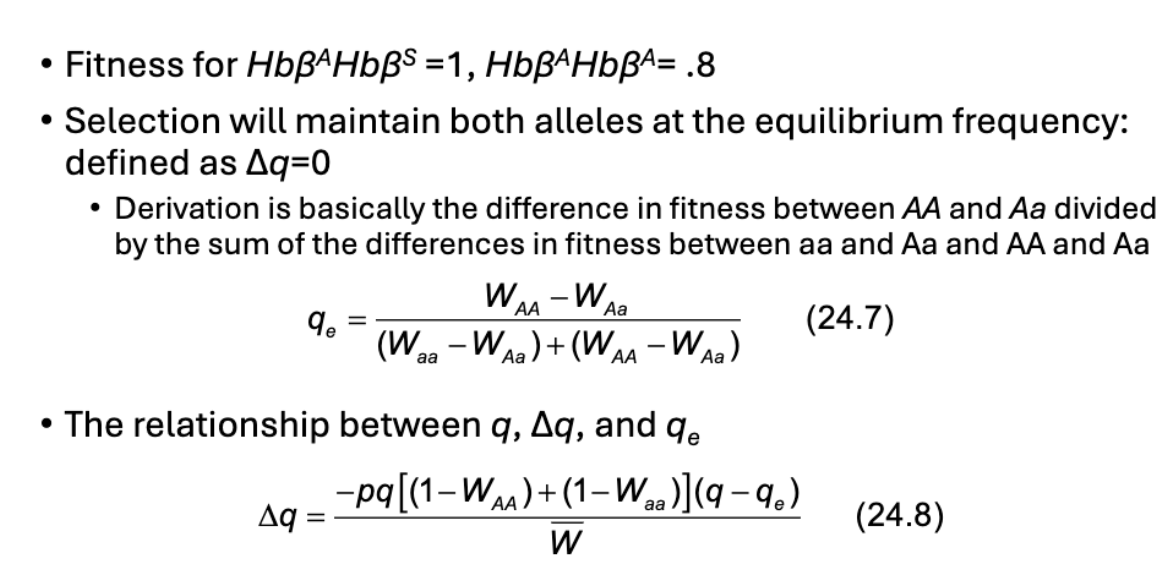
What is Sickle Cell and Malaria relations to Monte Carlo
Sickle cell anemia is recessive trait caused by mutations in B-globin locus
Heterozygous advantage: individuals that are carriers of sickel-cell are resistant to malaria
Sickle-cell allele distribution matches malaria distribution
What is balancing selection?
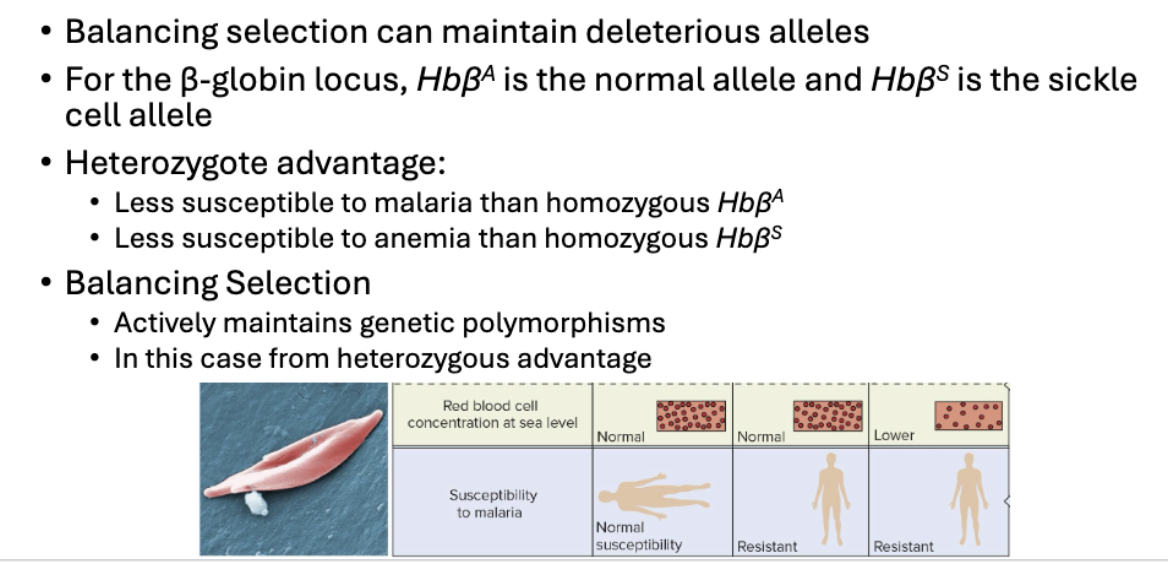
What’s the formula for balancing selection?
Natural Selection - Ex: Pesticide Resistance
Large-scale use of DDT and other synthetic insecticides began in 1940s
DDT is nerve toxin in insects
Dominant mutations in single gene confer resistance through detoxification of DDT
With insecticide application, strong selection favors heterozygotes
Resistance to every known insecticide has evolved with 10 years of its commercial introduction
Describe DDT resistance in mosquitos
Use of DDT Bangkok to control A. aegytpi mosquitos - began in 1964 and discontinued in 1967
R DOMINANT, RESISTANT, r is SUSPECTIBILITY
RR Genotype - Fitness cost
Absence of insecticide, resistance is subject to negative selection
What is Biological Ancestry?
Each person alive today has 2^k biological ancestors k generators ago, assuming ancestors were not related
What is Genetic Ancestry?
Inheritance of segments of DNA from biological ancestors
At each genetic locus there are only 2 genetic ancestors (one allele from each parent)
What is the Most Recent Common Ancestor (MRCA)
In this pedigree, the genetic pedigrees coalesce to one ancestral allele: the MRCA
Is MRCA is genome region specific?
YES.
Different genome regions have different MRCAs because of recombination
Exceptions: Y-Chromosome and mtDNA
Y-Chromosome don’t recombine and are paternal only
Mitochondrial genome is inherited maternally
How do Genetic Ancestry and mutations relate?
An unbroken line of descent connects the MRCA with all modern humans
The modern sequences are not identical, because mutations occur along the lineages
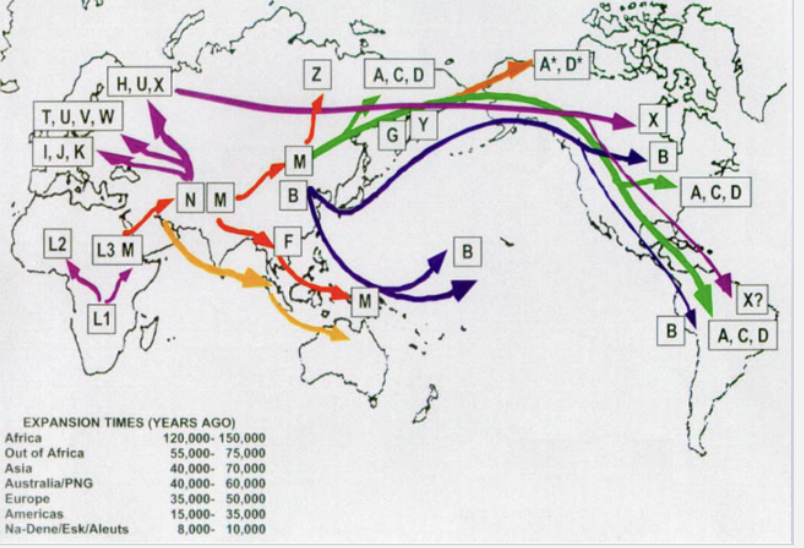
Where did Modern humans originate?
Most recent common ancestral mtDNA from mitochondrial Eve: lived no more than 200,000 years ago in Africa
Similarly, Y chromosome Adam lived 200,000 to 300,000 years ago in Africa
Did not interact and were likely separated by thousands of years
How do Mitochondrial Haplotypes work?
MITOCHODRIA DON’T RECOMBINE
Only new way is to generate new haplotypes
What is Mitochondrial Eve? What does Mitochondrial haplotypes provide?
By looking at mitochondrial haplotypes - can see how closely related various populations are
Estimating based on molecular clock and ancient DNA can provide information about when different events happened
How does Human Migration and mtDNA relate?
By plotting the mitochondrial haplotypes on a map along the information about when they diverged, we can trace migrations
Y Chromosomal Adam - What is this?
Y-Chromosome has the same trait of almost all change in haplotype from mutation
Lets us perform same experiment
Result is very similar
What are Founder Effects in H. sapiens migration
African populations have more DNA sequence diversity than other populations
As small populations left, founder effect reduced genetic diversity
What are some bottlenecks in human history?
Significant bottleneck occured 50,000 years ago in non-African populations
Estimated effective population size dipped below 2500
What are Neanderthal & Denisovan Genomes
Genomic DNA sequence was determined from bone samples
DNA sequence of certain genes allows predictions of appearance
What are some DNA Markers of inbreeding?
Various populations have markers found in Neanderthal and Denisovan genomes
Non-African humans have Neanderthal markers
Asian populations have Denisovan markers