🫷 Week 9 - Operons
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
43 Terms
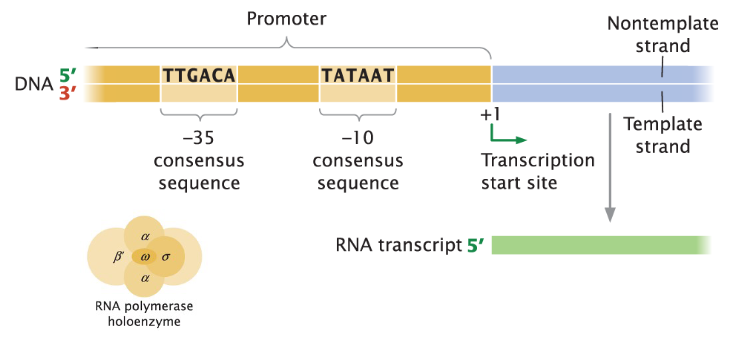
Transcription in Prokaryotes
Sigma Factor Function
Sigma factor (σ) recognizes and binds to the -35 and -10 consensus sequences in the promoter region
Positions RNA polymerase correctly to begin transcription
-10 Consensus Sequence
AT-rich, making it prone to unwinding
Facilitates the initiation of transcription
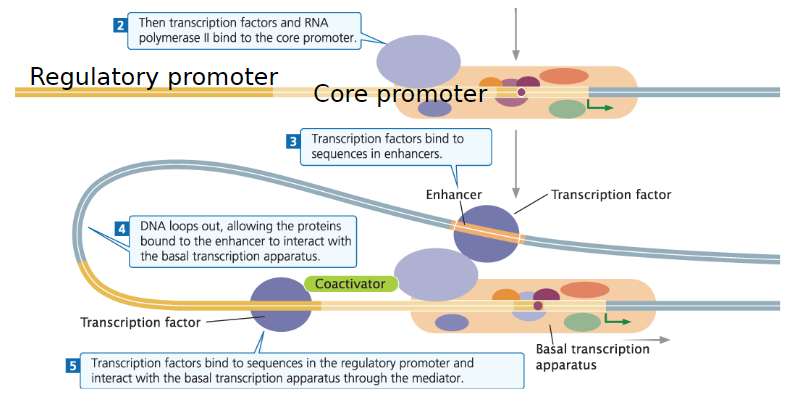
Transcription in Eukaryotes
Initiation
Assembly of transcription factors (TFs) and RNA polymerase II causes 11–15 bp of surrounding DNA to unwind
Template strand is positioned within the active site of RNA polymerase II
Promoter Structure
Each gene has a unique regulatory promoter
Contains distinct regulatory elements and unique cofactors to influence transcription
Regulatory promoter works with the core promoter to initiate transcription
Section 16.2 Outline: Gene Regulation
Topics Covered
1 Constitutive vs Inducible vs Repressible
2 Regulation Overview
3 Negative Inducible Operon
4 Negative Repressible Operon
5 lac Operon
6 Mutations
7 Positive Control and Catabolite Repression
Notes
Operator or promoter mutations and the trp operon will not be covered
Sections 16.3 and 16.4 will not be covered
Constitutive vs Inducible vs Repressible Genes
Constitutively Expressed Genes
Certain gene products, such as tRNAs, rRNAs, ribosomal proteins, RNA polymerase subunits, and enzymes for housekeeping functions, are essential in almost all cells
Genes for these products are continuously expressed in most cells (constitutive)
Inducible and Repressible Genes
Some gene products are needed only under specific environmental conditions
Inducible genes are expressed only when needed
Repressible genes are shut down when their products are no longer needed
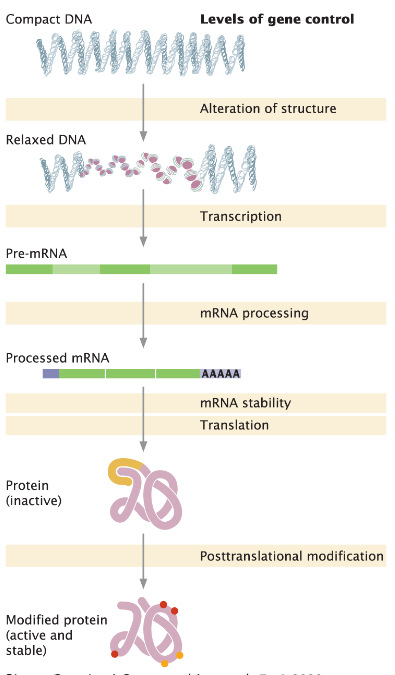
Regulation of Gene Expression
Regulation of Gene Expression
Gene expression can be controlled at multiple levels
Focus here is on regulation at the level of transcription initiation
Requires binding of proteins, called transcription factors, to the DNA
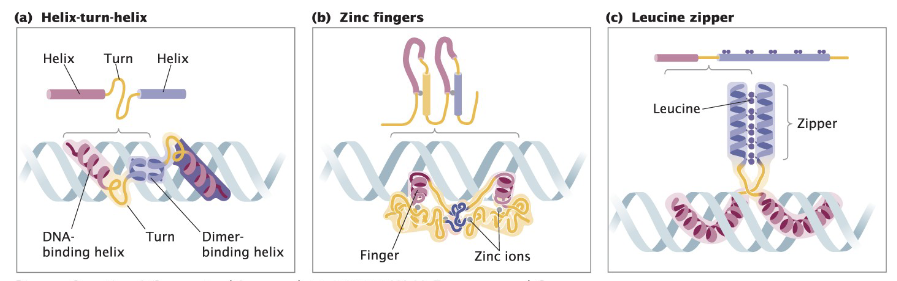
DNA Binding Proteins
DNA binding proteins can act as activators or repressors of transcription
Common in both prokaryotes and eukaryotes
Contain domains of 60–90 amino acids that recognize specific DNA sequences and interact with DNA grooves
Motifs bind non-covalently to the promoter of a gene or operon
Can recruit RNA polymerase or inhibit its binding
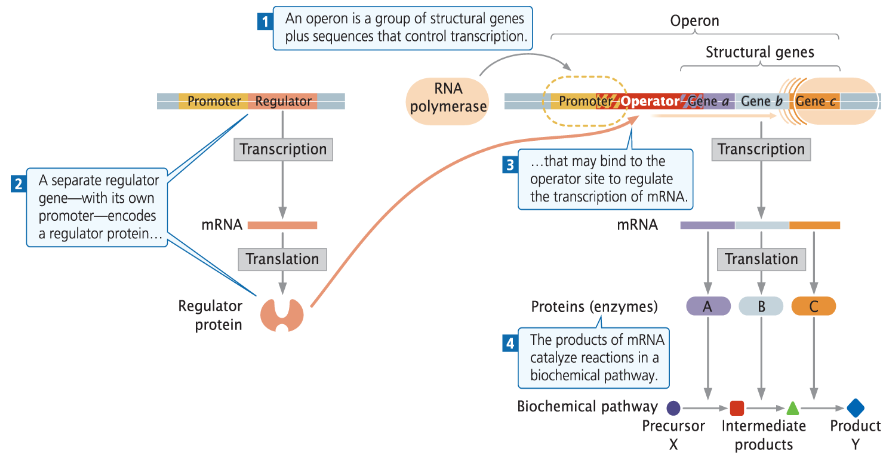
Transcription in Bacteria: Operons
Genes a, b, c are transcribed as a single mRNA
Transcription is regulated by a regulator protein, which is a transcription factor
Regulator binds to the operator part of the promoter
Operon Structure
An operon is a group of structural genes plus sequences that control transcription
A separate regulator gene with its own promoter encodes a regulator protein that may bind the operator site to regulate mRNA transcription
Function
mRNA is translated into proteins or enzymes
The products of mRNA catalyze reactions in a biochemical pathway
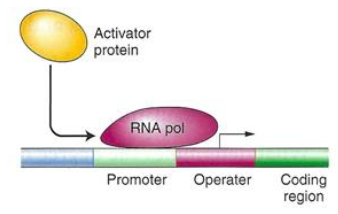
Control Elements in Bacterial Operons
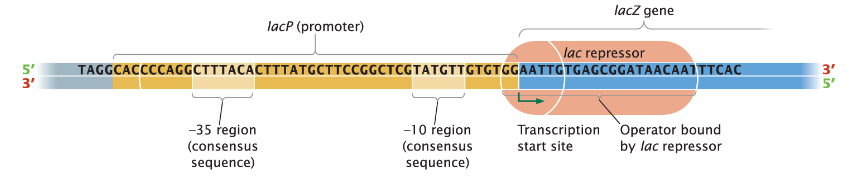
Operator
Part of the operon that helps determine whether transcription can take place
Overlaps with the 3' end of the promoter and the 5' end of the transcription start site of the first structural gene
Operator sequence is unique for each operon
Regulation of Transcription in Operons
Promoter/Operator Function
Regulates whether transcription of the operon occurs
Sometimes transcription needs to be turned on, other times it needs to be shut down
Regulatory Proteins
Negative regulatory proteins (repressors) inhibit transcription
Positive regulatory proteins (activators) stimulate transcription
Operon Types
Inducible operons are normally off and are turned on when needed
Repressible operons are normally on and are turned off when needed
Several varieties of control exist depending on individual cell needs
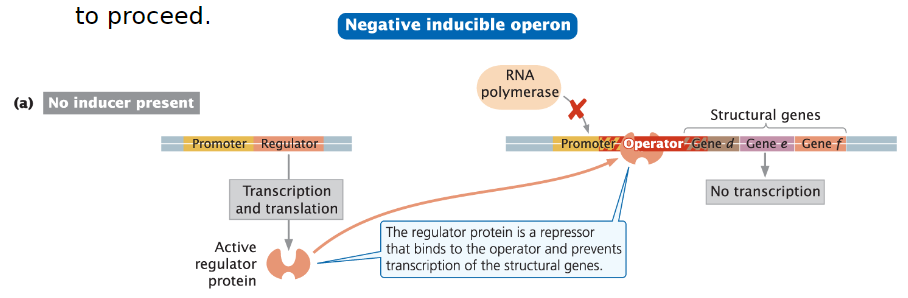
Negative Inducible Operon
General Function
Transcription is normally off and must be turned on
Regulator Protein
Regulator gene encodes an active repressor protein
Repressor binds to the operator and blocks RNA polymerase from binding to the promoter
Keeps transcription off
Repressor must be inactivated or removed for transcription to proceed
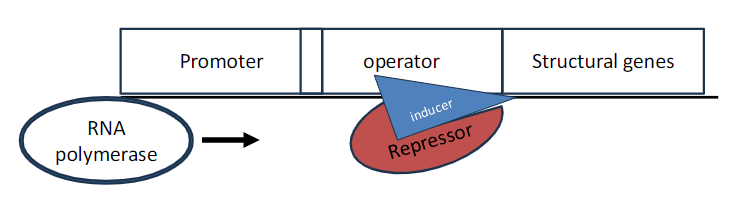
Negative Inducible Operon: Role of Inducer
Repressor Activity
Repressor is active in the absence of a co-factor
Binds to the operator and prevents transcription of structural genes
Inducer Function
A small molecule called an inducer binds to the repressor and inactivates it
Repressor can no longer bind to DNA
RNA polymerase can activate transcription
When the inducer is present, it binds to the regulator, making the regulator unable to bind to the operator
Transcription takes place
General Notes
Negative means the regulator protein is an inhibitor or repressor
Inducible means something inactivates the repressor, which then induces transcription
Usually involved in the degradation or metabolism of molecules
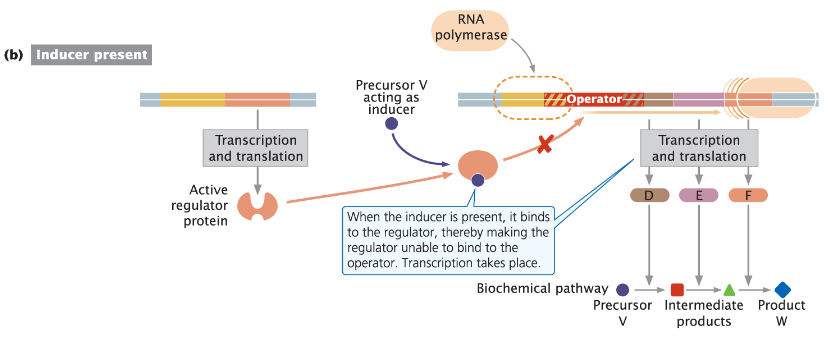
Negative Inducible Operon: Stepwise Mechanism
Without Inducer
Repressor protein binds to the operator region of the operon
Prevents transcription of the structural genes
Blocks RNA polymerase from accessing the promoter
With Inducer
Inducer binds to the repressor protein, causing conformational changes
Repressor can no longer bind to the operator
RNA polymerase can access the promoter and initiate transcription of the structural genes
Transcription takes place
Function
This mechanism ensures that the genes are only transcribed when the inducer is present
Often signals that the cell needs the products of those genes
Operon Components
Promoter
Operator
Structural genes
Image with Repressor
Image with Inducer
Example: lac Operon (Negative Inducible Operon)
Lactose is the inducer in the lac operon of Escherichia coli
In the absence of lactose, the repressor binds to the operator, blocking transcription
When lactose is present, it binds to the repressor and inactivates it
Repressor can no longer bind to the operator
Genes responsible for lactose metabolism are transcribed
Negative Repressible Operon
General Function
Transcription normally takes place and must be turned off
Regulator Protein
Regulator is a repressor, similar to negative inducible operons
Repressor is inactive in the absence of a co-factor
Unable to bind to the operator or promoter/operator complex
Transcription
Structural genes are transcribed until the repressor is activated by a co-factor
Inactive repressor allows transcription to proceed
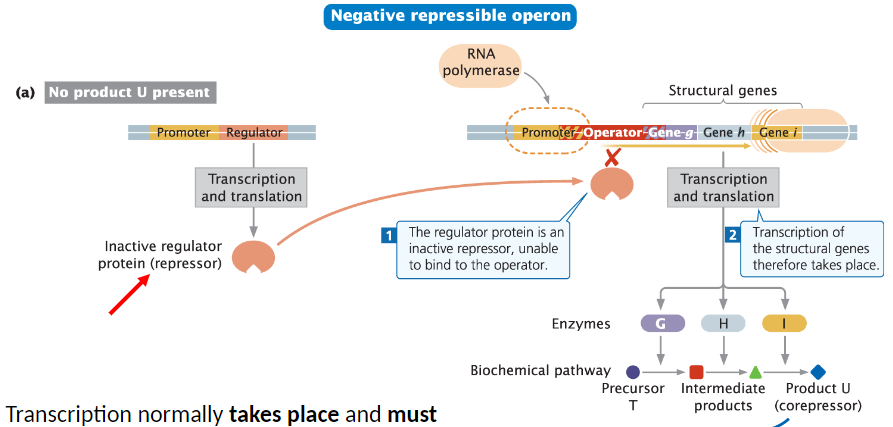
Negative Repressible Operon: Role of Corepressor
Corepressor Function
Small molecule called a corepressor (co-factor) binds to the repressor
Repressor/corepressor complex can now bind to the operator
Binding inhibits transcription
Function
Usually involved in biosynthesis necessary for the cell
Genes are only turned on when needed and off when not needed, making expression efficient
Negative means the regulator protein is an inhibitor or repressor
Repressible means something activates the repressor, which then represses transcription
Example: trp Operon
In Escherichia coli, product (e.g. tryptophan) builds up
Product binds to the regulator protein, making it active
Active repressor binds to the operator, preventing transcription
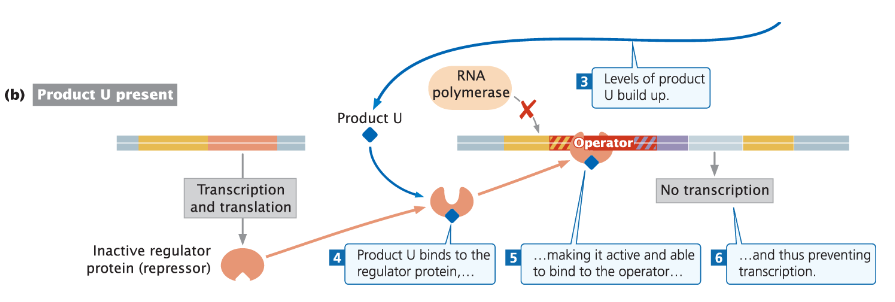
Example: trp Operon in E. coli
Repressor Protein
In its inactive form, the repressor cannot bind to the operator
Transcription of genes for tryptophan synthesis takes place
Co-repressor (Tryptophan)
When tryptophan levels are high, tryptophan acts as a co-repressor
Binds to the repressor protein
Transcription Inhibition
Binding of tryptophan to the repressor causes a conformational change
Active repressor binds to the operator and blocks transcription
Function
Ensures genes for tryptophan synthesis are transcribed only when tryptophan levels are low
Prevents the cell from wasting energy producing tryptophan when it is abundant
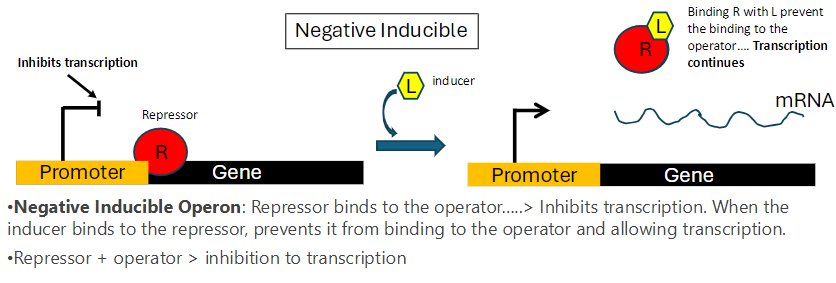
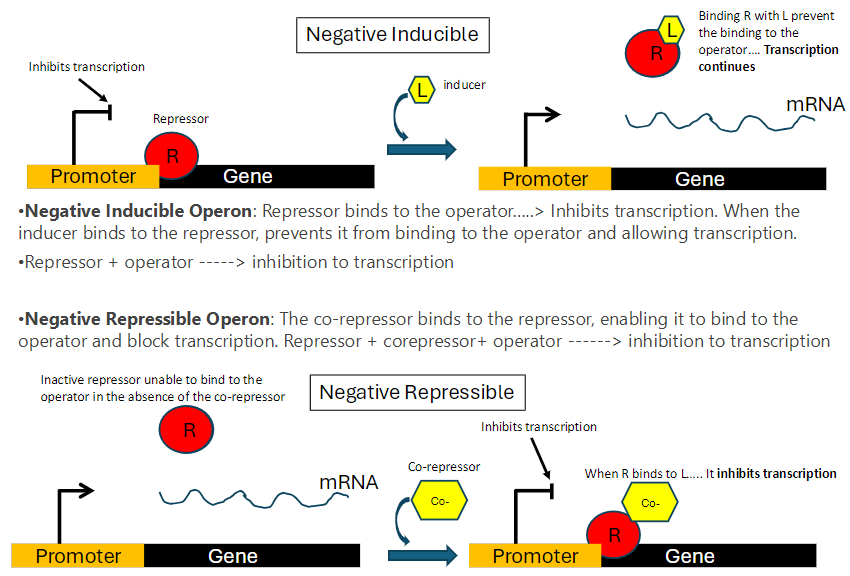
Negative Inducible Operon: Summary
Repressor Function
Repressor binds to the operator and inhibits transcription
Inducer Role
Inducer (L) binds to the repressor (R)
Binding prevents the repressor from attaching to the operator
Transcription continues
Mechanism
Repressor + operator = transcription inhibited
Repressor + inducer = repressor cannot bind operator, transcription proceeds
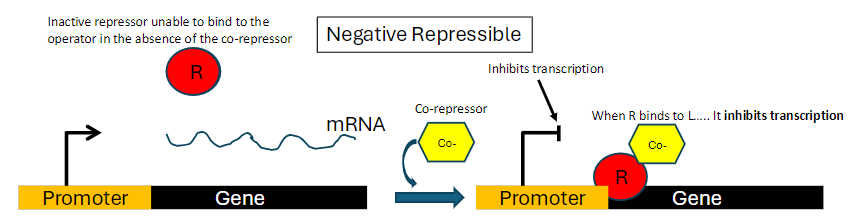
Negative Repressible Operon: Summary
Repressor Activity
Repressor is inactive and cannot bind to the operator in the absence of a co-repressor
Co-repressor Role
When co-repressor binds to the repressor, the repressor becomes active
Active repressor binds to the operator and inhibits transcription
Mechanism
Inactive repressor alone = transcription proceeds
Repressor + co-repressor = transcription inhibited
Negative Inducible vs Negative Repressible Operons
Negative Inducible Operon
Repressor is active by default and binds to the operator
Blocks transcription of structural genes
Inducer binds to the repressor, inactivating it
Repressor can no longer bind the operator
Transcription occurs
Typically involved in degradation or metabolism of molecules
Negative Repressible Operon
Repressor is inactive by default and cannot bind the operator
Transcription of structural genes normally takes place
Co-repressor binds to the repressor, activating it
Active repressor binds to the operator
Transcription is inhibited
Typically involved in biosynthesis of molecules
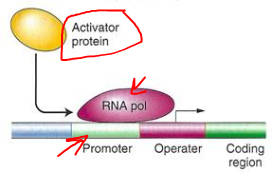
Positive Control of Transcription
Activator Proteins
Regulatory protein acts as an activator
Activator binds to the operator or upstream of the operator to induce transcription
Catabolite Activator Protein (CAP)
CAP is a positive activator of transcription
Binds just upstream of the promoter
Enhances binding of RNA polymerase to the promoter
Lac Operon Regulation
Negative Inducible Control
Regulator protein is an inhibitor
Allolactose acts as an inducer by inactivating the inhibitor
Positive Control
CAP combined with cAMP enhances transcription
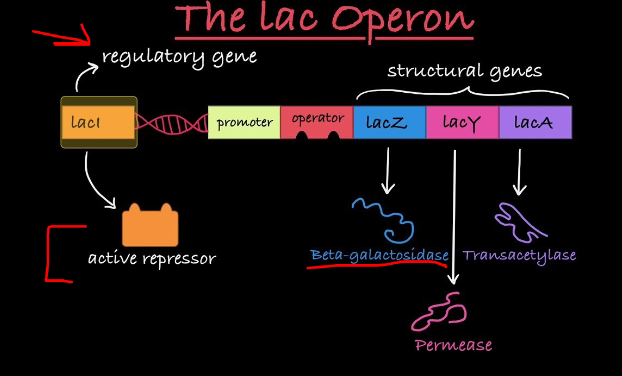
Lac Operon Enzymes and Function
Lactose Metabolism
Lactose, found in milk, can be metabolized by E. coli
Needs to be transported into the cell by permease (lacY)
β-galactosidase (lacZ) breaks lactose into glucose and galactose
β-galactosidase also converts lactose into allolactose
Thiogalactoside transacetylase (lacA) is the third enzyme, function unclear
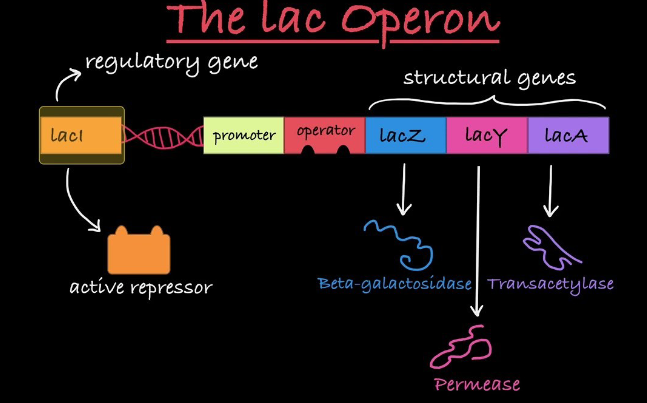
Operon Structure
All enzymes are encoded by adjacent structural genes
They share a common promoter (lacP)
Function Summary
Permease actively transports lactose across the cell membrane
β-galactosidase breaks lactose into galactose and glucose
β-galactosidase also converts some lactose into allolactose, which acts as the inducer
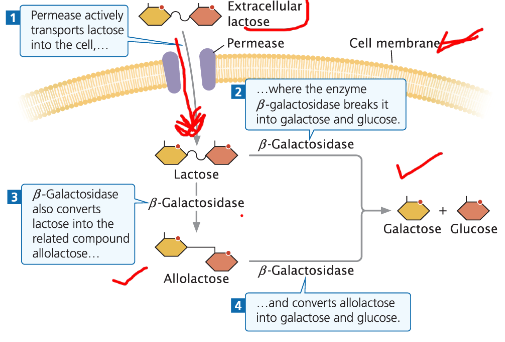
Lac Operon Induction
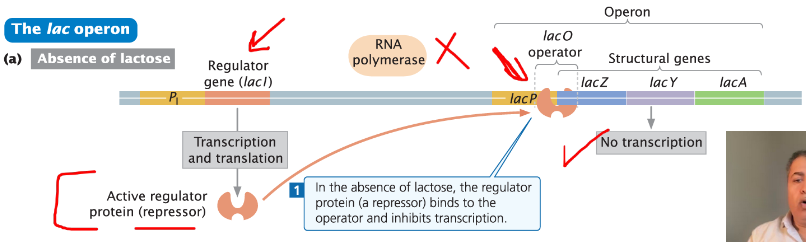
Absence of Lactose
Very little transcription of the operon occurs, but not none
Repressor protein (lacI) binds to the operator (lacO) and inhibits transcription
Presence of Lactose
Addition of lactose to the medium instead of glucose increases transcription of the lac operon about 1000X within 2–3 minutes
Lactose acts as the inducer
Coordinate Induction
Simultaneous synthesis of several proteins by a specific inducer molecule
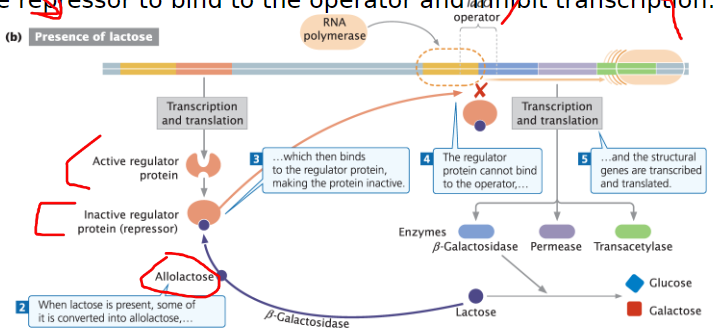
Lac Operon: Mechanism When Lactose is Present
Allolactose Formation
Lactose is converted into glucose, galactose, and some allolactose
Allolactose keeps the operon in the “on” position
Repressor Inactivation
Allolactose binds to the repressor, causing it to release from the operator
Repressor cannot bind to the operator
Transcription and Translation
RNA polymerase binds to the promoter and initiates transcription of lacZ, lacY, and lacA
Structural genes are transcribed and translated
Termination
Once lactose is depleted, no more allolactose is produced
Repressor binds again to the operator, inhibiting transcription
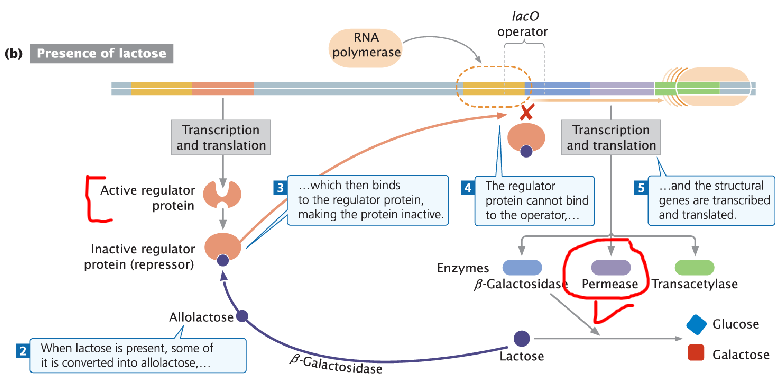
Lac Operon: Initial Activation
Low-Level Transcription
Repression by the repressor does not completely shut down transcription
Low levels of transcription occur even when the repressor is bound
Low Enzyme Levels
Low levels of permease (lacY) and β-galactosidase (lacZ) are always present in cells
These basal amounts allow lactose to enter the cell and be converted into allolactose to inactivate the repressor
Lac Operon in Escherichia coli
Repressor Protein
In the absence of lactose, the repressor binds to the operator
Prevents transcription of genes responsible for lactose metabolism
Inducer Molecule (Allolactose)
When lactose is present, it is converted into allolactose
Allolactose binds to the repressor and causes a conformational change
Transcription Initiation
Repressor can no longer bind to the operator
RNA polymerase accesses the promoter and transcribes the genes needed for lactose metabolism
Function
Ensures genes are transcribed only when lactose is available
Allows the cell to efficiently utilize lactose as an energy source
Lac Operon: Stepwise Mechanism
Step 1 – Basal Transcription
Low-level transcription occurs even when the repressor is bound
Small amounts of permease (lacY) and β-galactosidase (lacZ) are present
Step 2 – Lactose Entry
Lactose enters the cell through basal levels of permease
Step 3 – Inducer Formation
Lactose is converted into allolactose by β-galactosidase
Allolactose acts as the inducer
Step 4 – Repressor Inactivation
Allolactose binds to the repressor, causing a conformational change
Repressor can no longer bind to the operator
Step 5 – Transcription Initiation
RNA polymerase binds to the promoter
Structural genes lacZ, lacY, and lacA are transcribed
Step 6 – Translation and Function
mRNA is translated into enzymes
Permease imports more lactose, β-galactosidase breaks lactose into glucose and galactose
Allolactose production continues, keeping the operon “on”
Step 7 – Termination
When lactose is depleted, no allolactose is made
Repressor binds to the operator again
Transcription is inhibited
Discovery of Gene Regulation: Jacob and Monod
Key Findings
Genes can be turned on or off depending on environmental conditions
A group of genes in the same metabolic pathway can be controlled together by a single regulatory system called an operon
Operon Components
Structural genes code for enzymes
Promoter is where RNA polymerase binds
Operator is a DNA segment that acts as an on/off switch
Repressor protein binds to the operator to block transcription when the gene is not needed
Mutations
Studying the lac operon, they discovered mutations in certain genes that can alter how the operon is regulated
Mutations Affecting Lac Operon Regulation
lacI⁻ Mutations
Occur in the repressor gene
Prevent production of a functional repressor
Operon is always on, even without lactose
lacOᶜ Mutations
Occur in the operator sequence
Prevent repressor from binding
Operon is continuously expressed
Significance
Specific mutations can alter control of gene expression
Reveals how DNA sequences and regulatory proteins interact
Explains how mutations in regulatory genes or DNA sites disrupt operon control, a key insight in molecular genetics
Mutations Affecting Lac Operon Proteins
LacZ and LacY Mutations
Mutations in LacZ or LacY disrupt lactose metabolism
Affect the function of the proteins
Do not affect the regulation of their synthesis
Studying Gene Regulation with Mutations
Experimental Approach
Examined single gene mutations in E. coli
Crossed mutant cells via conjugation
Purpose
Applied different combinations of mutations in plasmids or chromosomes
Determined how the gene was regulated under different genetic conditions
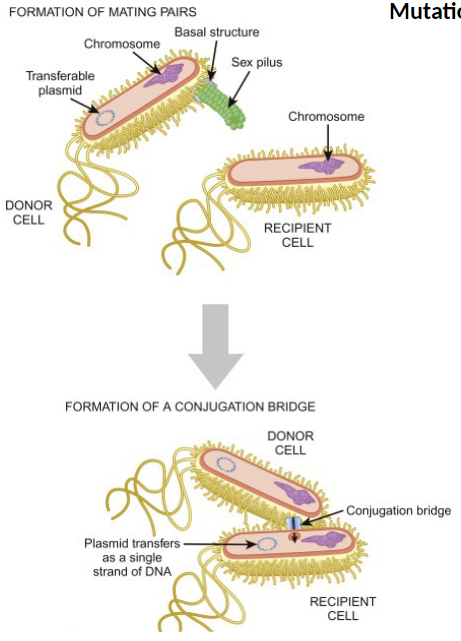
Cis and Trans Regulation of Gene Expression
Cis Regulation
Controls gene expression only on the same piece of DNA
Example: promoter sequence of the lac operon acts in cis on lacZ
Trans Regulation
Controls gene expression on other DNA molecules
Example: repressor protein acts in trans on lacZ
Partial Diploids
Bacteria are typically haploid
A plasmid carrying the lac operon is added, creating a partial diploid
Not every gene is in the plasmid, allowing study of cis and trans effects
Cis Regulation
Cis = DNA part
It only controls the gene right next to it
It cannot move
Example:
The operator sequence
If it’s broken, only that operon is affected
Trans Regulation
Trans = protein
It can move around the cell
It can control any copy of the gene
Example:
The lac repressor protein (LacI)
It can bind to operators on ANY DNA
Partial Diploids
Bacteria get a plasmid with another lac operon
Now you can see:
If the mutation only affects its own operon → cis
If the mutation affects both operons → trans
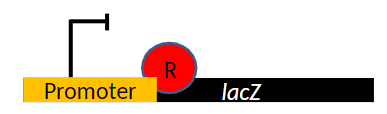
Notation for Mutations
Mutation Symbols
Non-functional mutations are designated with a “-” superscript, e.g. lacZ-
Wild type is designated with a “+” superscript, e.g. lacZ+
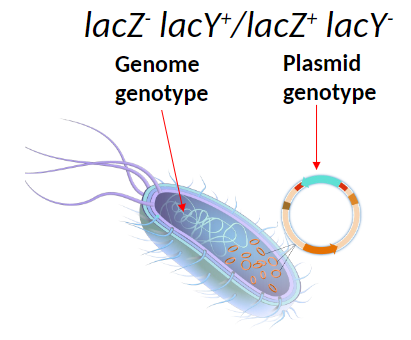
Partial Diploid Example
Bacteria have a mutation in the lacZ gene and wild type lacY in the genome
Plasmid carries wild type lacZ and mutant lacY
Genome genotype: lacZ- lacY+
Plasmid genotype: lacZ+ lacY-
One good copy of each gene allows the bacterium to metabolize lactose
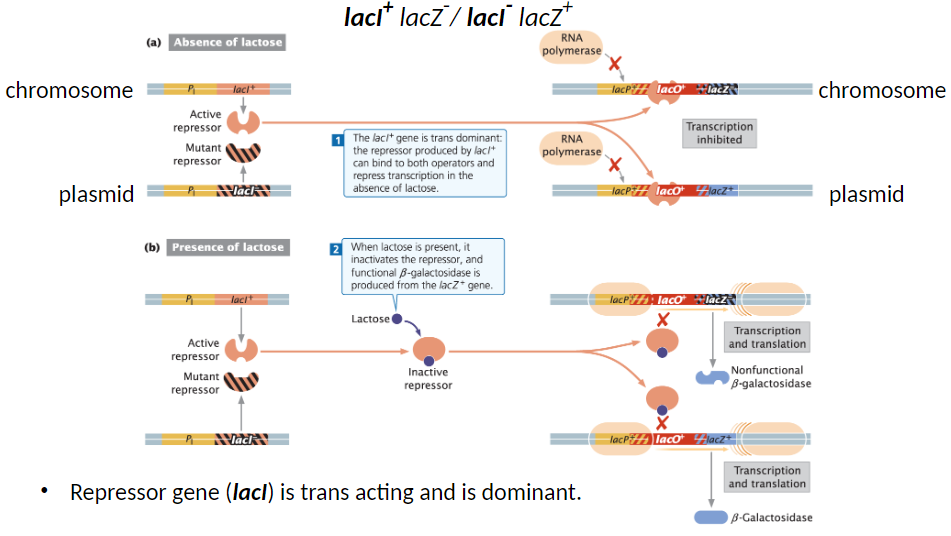
Trans Dominance of lacI
Partial Diploid Example
Genotypes: lacI+ lacZ- / lacI- lacZ+
lacI+ on one DNA molecule, lacI- on the other
Repressor Function
Repressor gene (lacI) is trans acting and dominant
Repressor produced by lacI+ can bind to both operators and repress transcription when lactose is absent
Lactose Effect
When lactose is present, it inactivates the repressor
Functional β-galactosidase is produced from lacZ
Bacteria functions normally, like wild type, in the presence or absence of lactose
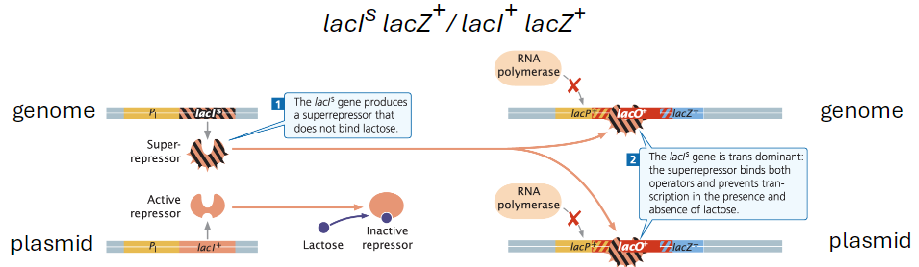
Super-Repressor Mutation (lacIs)
Partial Diploid Example
Genotypes: lacIs lacZ+ / lacI+ lacZ+
Super-repressor mutation makes repressor insensitive to allolactose
Repressor Function
Mutant repressor binds the operator in the presence or absence of allolactose
Called a super-repressor because it is very dominant
Blocks transcription regardless of lactose availability
Effect on Lactose Metabolism
Bacteria cannot metabolize lactose
Transcription of structural genes is inhibited independent of lactose
Super-repressor binds both operators and prevents RNA polymerase from initiating transcription
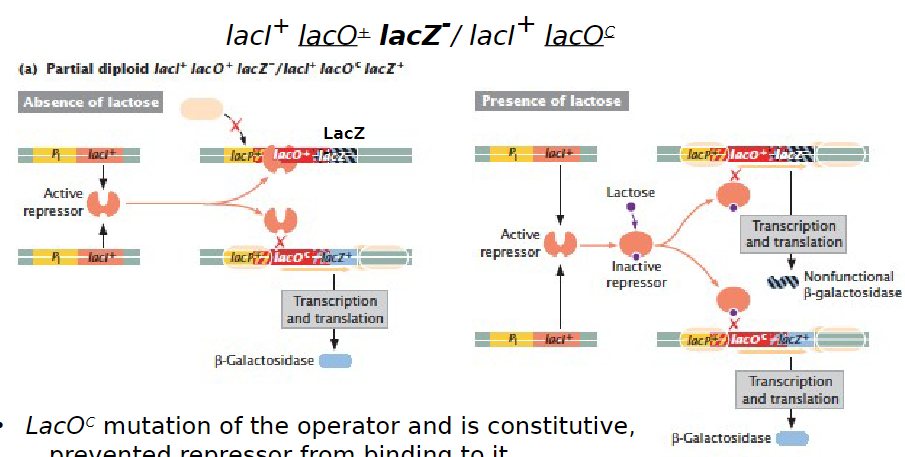
Constitutive Operator Mutation (lacOC)
Partial Diploid Example
Genotypes: lacI+ lacO+ lacZ- / lacI+ lacOC lacZ+
Operator Function
lacOC mutation prevents repressor from binding
lacO is cis acting, so only affects the genes on the same DNA molecule
Effect on β-Galactosidase Production
β-galactosidase is produced regardless of lactose presence when paired with lacZ+ on the same DNA
If lacZ+ were paired with lacO+ on the same sequence, production would occur only in the presence of lactose
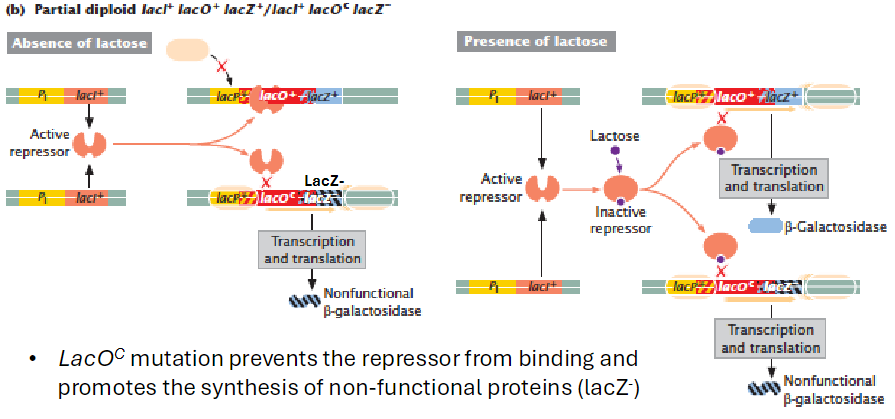
Effect of lacOC with Mutant lacZ
Partial Diploid Example
Genotypes: lacI+ lacO+ lacZ+ / lacI+ lacOC lacZ-
Operator and Protein Function
lacOC prevents repressor from binding
Promotes transcription of structural genes on the same DNA
Effect on β-Galactosidase
With wild-type lacZ, β-galactosidase is produced normally only when lactose is present
With mutant lacZ-, transcription produces non-functional protein in the presence or absence of lactose
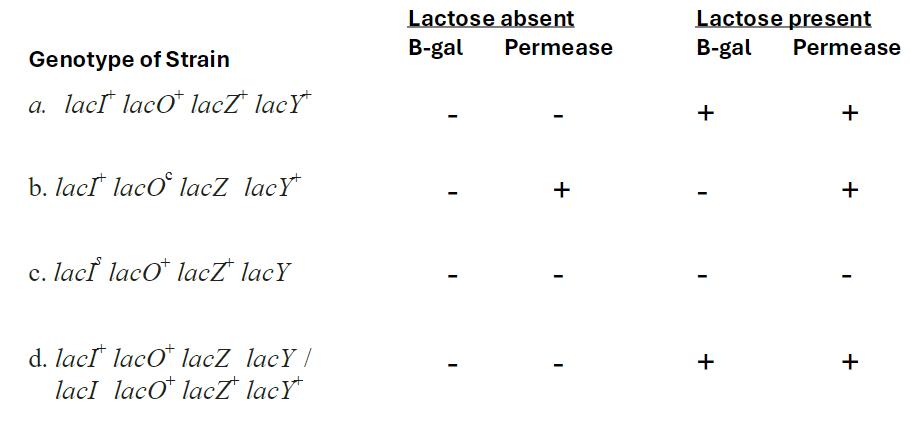
Lac Operon Expression Table
Strain a: lacI lacO lacZ lacY
Lactose absent: B-gal +, Permease -
Lactose present: B-gal +, Permease +
Strain b: lacI+ lacO lacZ lacY
Lactose absent: B-gal +, Permease -
Lactose present: B-gal +, Permease +
Strain c: lacI lacO lacZ lacY
Lactose absent: B-gal -, Permease -
Lactose present: B-gal +, Permease +
Strain d: lacI lacO lacZ lacY / lacI lacO lacZ lacY
Lactose absent: B-gal +, Permease +
Lactose present: B-gal +, Permease +
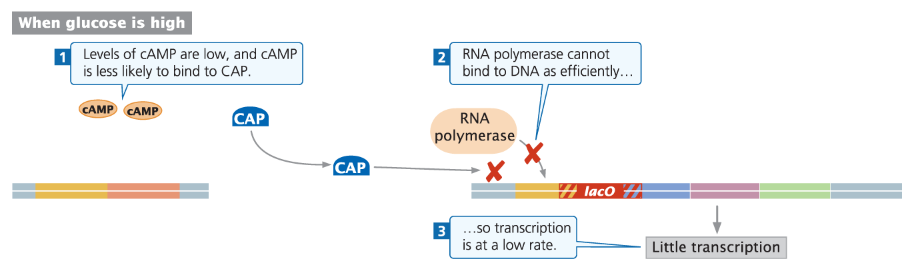
Positive Control and Catabolite Repression
Glucose Preference
Glucose is the preferred energy source because it requires less energy to metabolize
Bacteria preferentially metabolize glucose even when other sugars like lactose are present
Catabolite Repression
When glucose is present, bacteria turn off other metabolic pathways
This repression of alternative pathways is called catabolite repression
Example: glucose metabolism represses the lac operon
Positive Control
When glucose is low, transcription of other metabolic pathways is turned on
This positive control is independent of the repressor/inhibitor mechanisms
Molecular Mechanism
Positive control works via catabolite activator protein (CAP) and cAMP
Levels of cAMP are inversely proportional to glucose levels
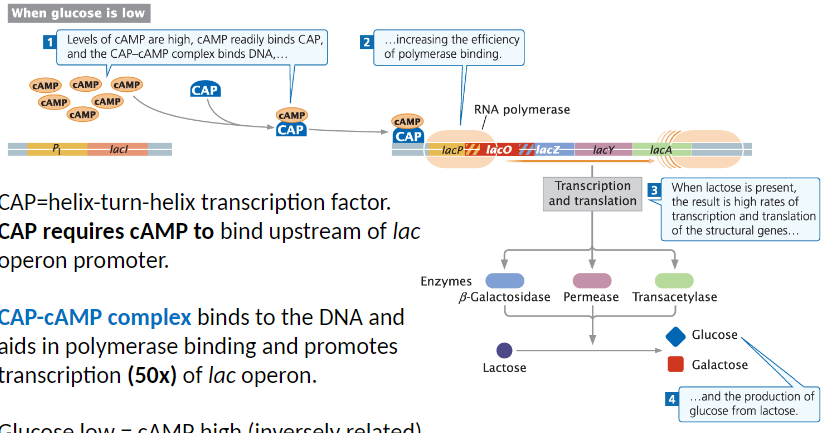
CAP and Positive Control of the Lac Operon
CAP Function
CAP is a helix-turn-helix transcription factor
Requires cAMP to bind upstream of the lac operon promoter
CAP-cAMP Complex
CAP-cAMP binds DNA and aids RNA polymerase binding
Promotes transcription of the lac operon up to 50x
Glucose and cAMP Relationship
Glucose low → cAMP high
CAP-cAMP exerts positive control on more than 20 operons in E. coli
Effect on Lac Operon
When glucose is low and lactose is present, CAP-cAMP binds DNA
Increases RNA polymerase efficiency
Results in high rates of transcription and translation of structural genes
Leads to production of glucose from lactose
Effect of High Glucose on Lac Operon
cAMP Levels
High glucose → cAMP levels are very low
CAP-cAMP Complex
Few or no CAP-cAMP complexes form
Reduced binding of RNA polymerase to lac promoter
Expression Outcome
Significantly reduced transcription of lac operon
Lactose metabolism is minimized while glucose is present