UBC biol 203
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
42 Terms
What is a Protist?
Eukaryotes that aren't animals, plants, or fungi
Leeuwenhoek
Discovered the first single cellular organism (cork).
Haeckel
added to biodiversity, protists on top of animals and plants
Whittaker
5 kingdoms, protista & monera (no higher/lower organisms)
Woese
discovered archaea through phylogentic taxonomy of small subunit rRNA
Posterior Probability
probability that the tree is correct
Bootstrap %
confidence for each clade of a tree
Operational Taxonomic units
% similarity
monophyly
share a common ancester
polyphyly
grouped together by common features but not really related (convergent evolution)
Paraphyletic
do not include all descendents of a single common ancester
Phylogeny
Phylogentic tree, hypothesis about evolutionary relationships
How did eukaryote complexity come to occur?
endosymbiosis
Bacteria
2 membrane
peptoglycan membrane, gram negative
flagella (secretion, signalling, sensory)
key to success: metabolism
Eukaryote
plasma membrane
cytoskeleton: tubulin & actin
endomembrane system: transport lipids, protiens, ER, nucleus, golgi, vesicles
Difference in transcription in bacteria and eukaryotes
Bacteria: RNA polymerase binds to -35 -10 box with sigma factor, Polycistronic mRNA (can encode for more than one protiens), operons
Eukaryotes: TATA binding protien binds to TATA box with RNA polymerase and Transcriptional factors. The resulting mRNA has a cap and a poly A tail.
Difference in translation in bacteria and eukaryotes
Bacteria: small subunit with tRNA (fmet) binds to shine dalgarno sequence, then large subunit comes to begin translation.
Eukaryotes: Cap binding protien binds to cap, small subunit with tRNA (met) is joined by large subunit to begin translation. Introns spliced out.
outgroup
more distant to org of interest than they are to each other
Iwabe
used translation elongation factors genome sequence to trace gene to LUCA, thus determining bacteria as the outgroup
Lokiarchaeota
has actin, cytoskeletal genes and endomembrane system which are fundamentals of eukaryotes
Archaea
extreme environment dwellers
have lipid membrane (saturated)
Transcription & translation like eukaryotes at molecular level but simpler
Endosymbiotic theory
Ancestors of mitochondria and plastids was prokaryotes that came to live in a host cell. supported by the fact that mitochondria and chloroplasts have DNA
Archezoa hypothesis
a kingdom proposed for eukaryotes that diverged before origin of mitochondria. Proven false when genetic residue of the mitochondrion is found in amitochondriates (protists lacking mitochondria)
Metamonads (diplomonads)
mostly parasites
tetrakont arrangement
giardia is possible link to mitochondria
mirror image cell (2 cells fused)
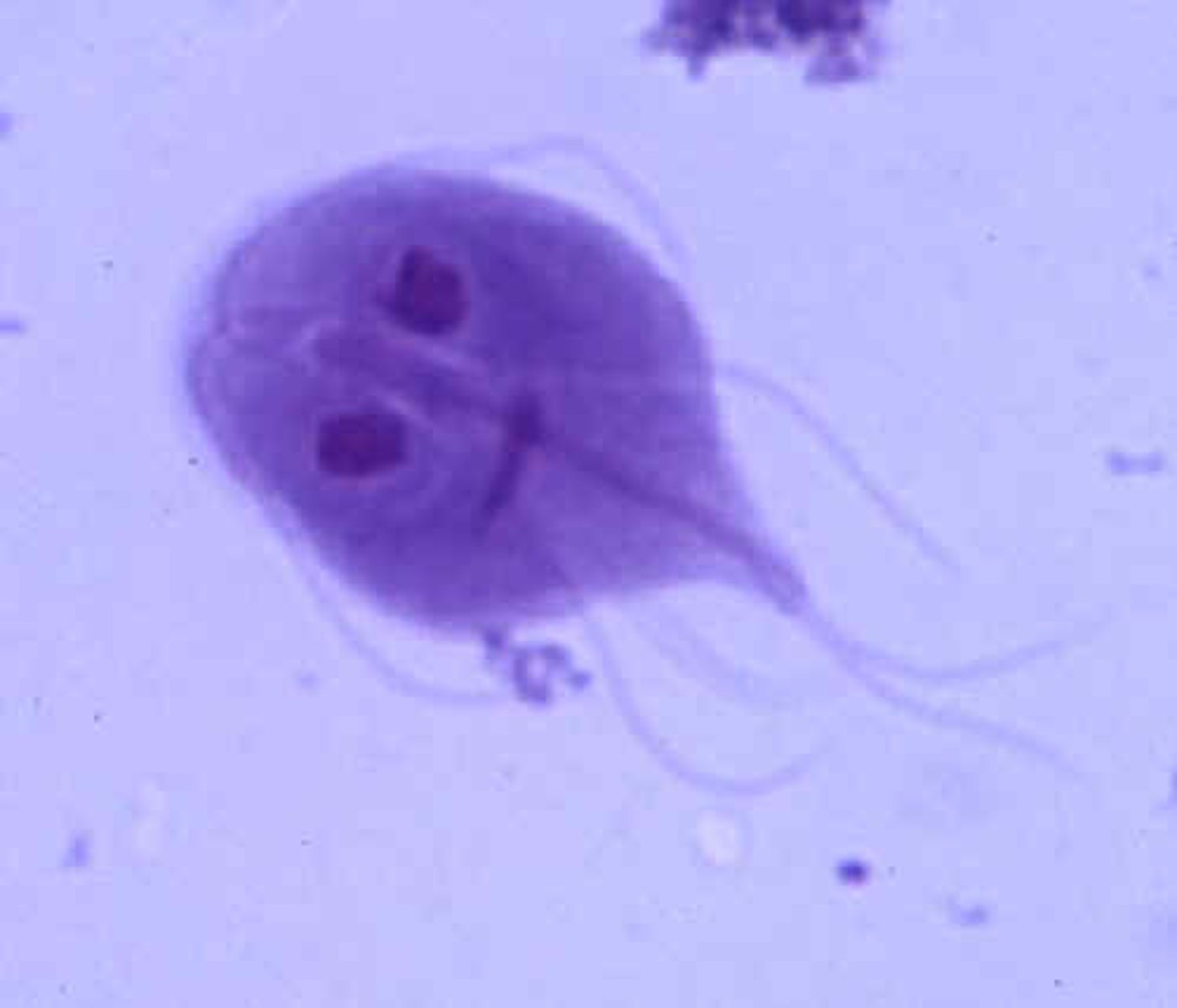
Parabasalia
mostly parasites, symbionts
tetrakont
recurrent flagellum
undulating membrane
costa (contracts)
axostyle
Hydrogenosome
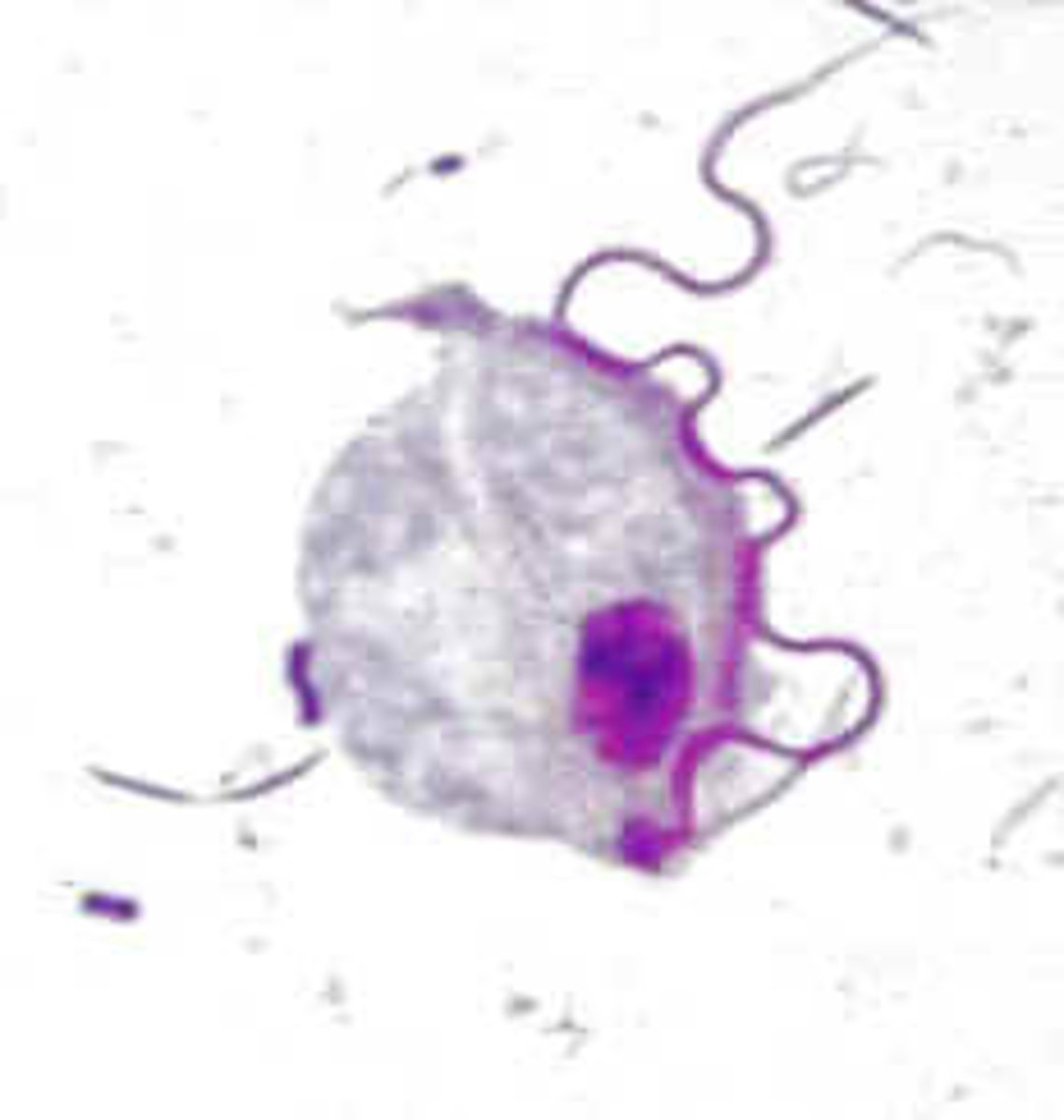
Hydrogenosome
no Oxidative phosphorylation
no electron transport
no cristae
no genome
not efficient but anoerobic
possible link to mitochondria
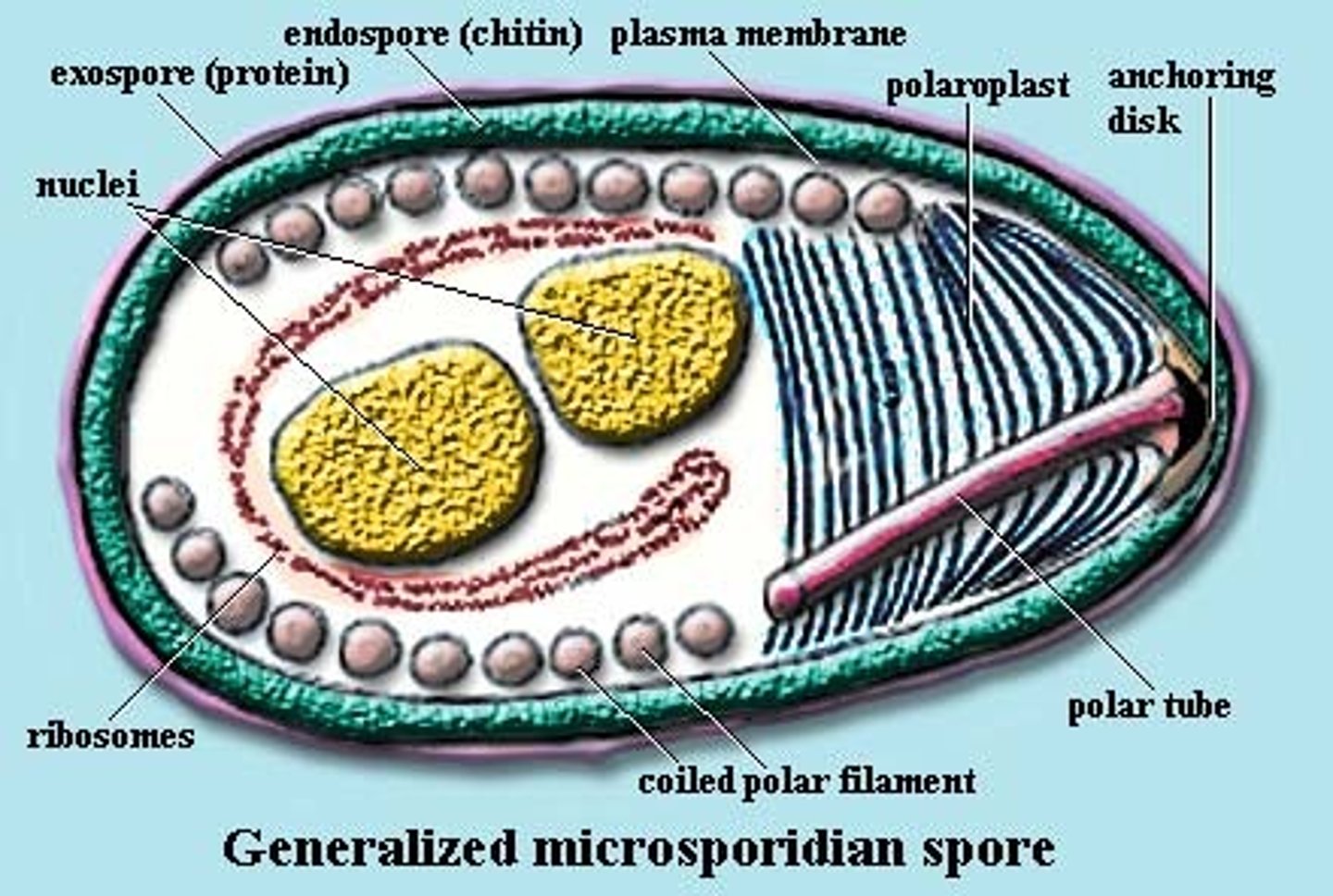
Microsporidia
all parasites (obligate intracellular)
spore (dormant)
Imports ATP
anaerobes
smallest nuclear genome
closely related to fungi (eukaryote)
anchoring disk
polar tube
two nucleus
dipplokaryon
posterior vacuole
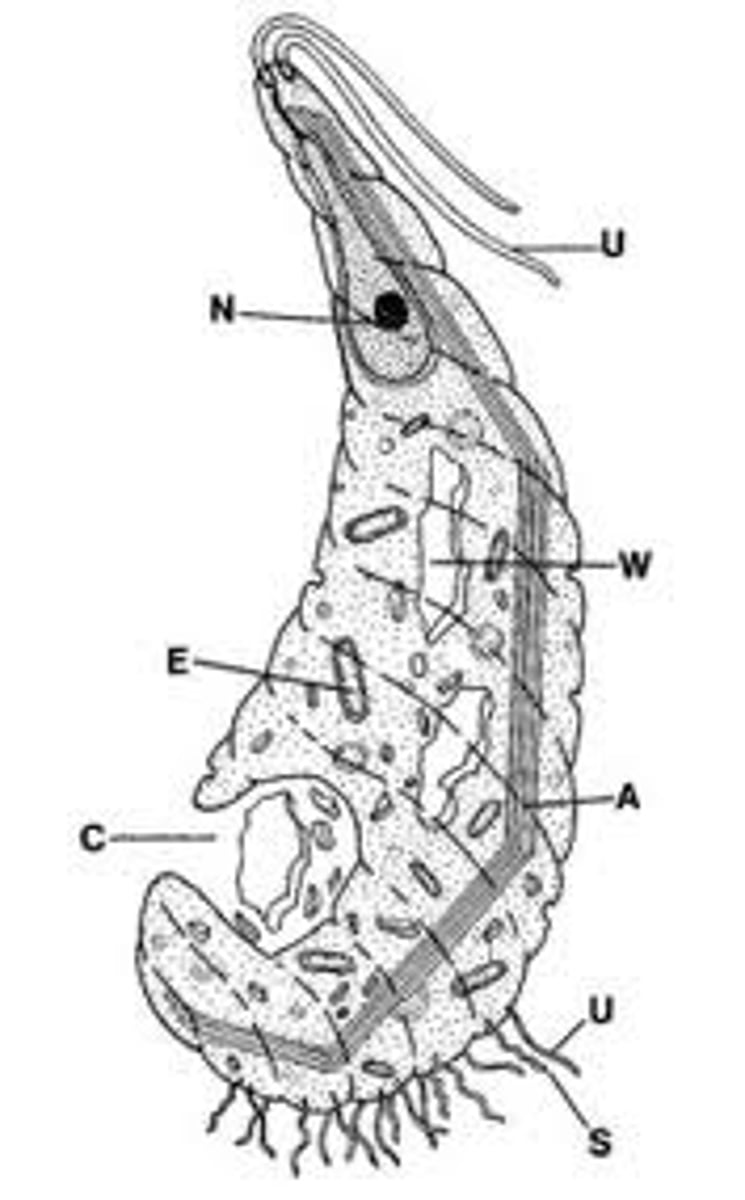
Oxymonad
all symbionts
anaerobic
to mitochondira/ organelles
no genes
Fe S cluster by horizontal transfer
LOST (only know eukaryote bc tree)
motile axostyle
pinocytic pore
mitosome/hydrogenosome (only FeS)
Archamoebae
some parasite & free living
branch near mitochondrial origin
evolved from mitochondrial containing ancesters
mitochondria
original gene from alpha-proteobacteria
makes ATP
Translation/Transcription DNA replication
Heme synthesis
FeS cluster
Cristae
Matrix
small and few genome
ATP production mitochondria
glycolysis: 2 ATP
Krebs cycle: 2ATP
Electron transport chain: 26ATP
Endosymbiotic gene transfer
genes present in organelle moved to nucleus but protiens are re-imported
Translocation outer membrane (TOM)
Translocation inner membrane (TIM)
Transit peptide
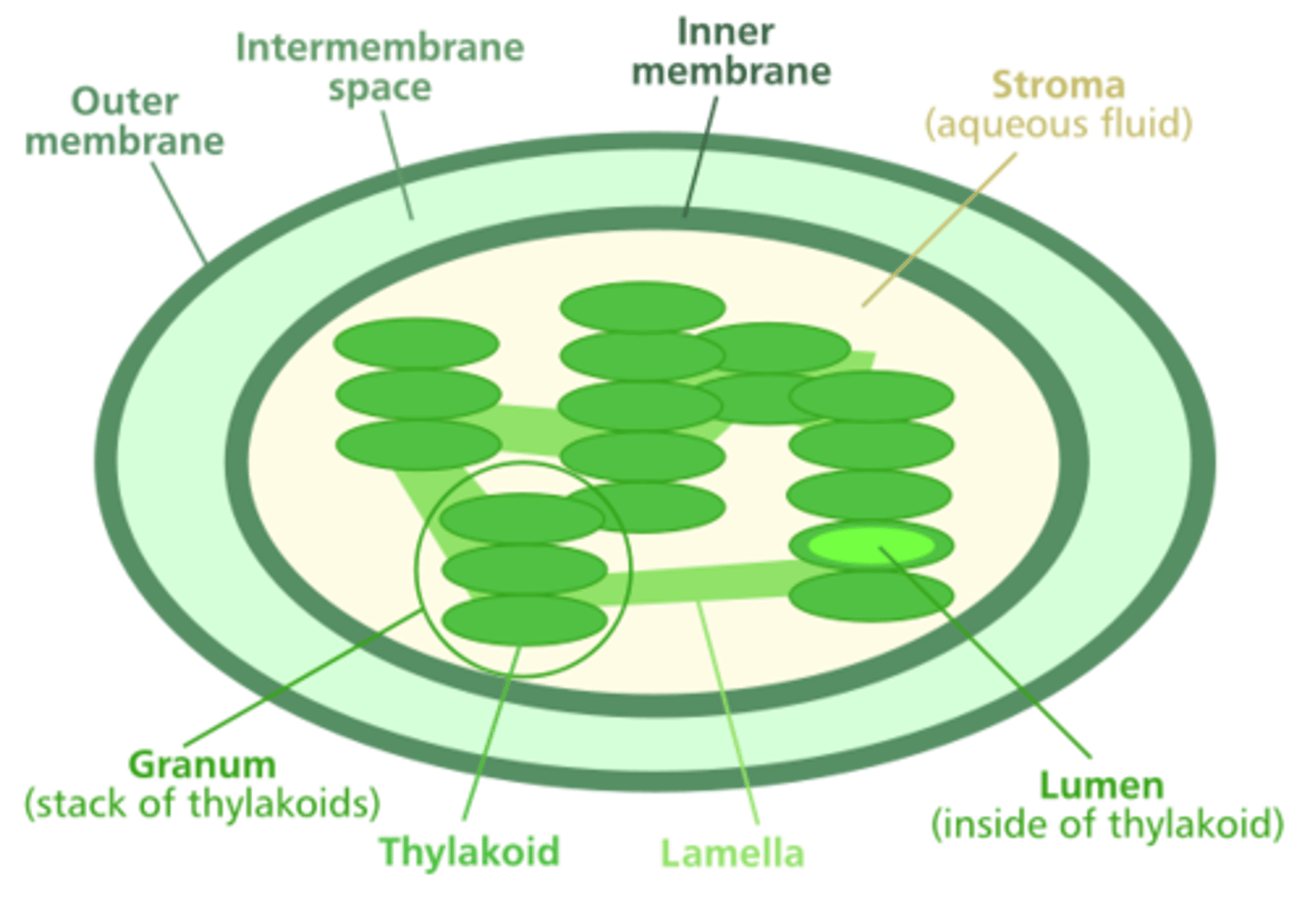
Plastids
evolved later (not all euk have plastids)
diverse
choloroplast
very closely related to cyanobacteria
photosynthesize and Biosynthesis
TOC & TIC
primary endosymbiosis
cyanobacteria + eukaryote = primary plastid (primary algae)
Green algae
Red algae
Glaucophyte (rare, never secondary endosymbiosis)
secondary endosysmbiosis
primary plastid + some other eukaryote = secondary plastid (secondary algae)
still retain outer membrane
contribute to protist diversity
Chromalveolate hypothesis
endosymbiosis of a red algae gave rise to eukaryotes
Nucleomorphs
contain reduced genone through gene loss and gene transfer
lost in all other algae with plastids of secondary endosymbiosis except Crytomonads and chlorarchinophytes
key evidence for secondary endosysmbiosis
secondary plastid protien secretion
signal peptide and transit peptide
post and co-translational target
directed to ER by Signal recognition particle, then TIC/TOC
transport to more than one cellular compartment
SELMA
symbiont-derived ERAD like machinery
diverts ERAD protein transport capabilites to mediate protein import to the second outmost plastid membrane
ERAD
ER associated degredation system
recycling misfolded proteins from ER
periplastidal compartment
PPC
nuclomorph in the 4 membrane
tertiary endosymbiosis
doesn't contribute to biodiversity
host already a dinoflagellate
non photosynthesis (lost it then re-gained)
symbiont relationship with diatom, haptophytes, cryptophytes
function as two individual cells