lec 1.3 - review of DNA and RNA
1/27
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
DNA/RNA nucleotide composition
sugar molecule - ribose (RNA) or deoxyribose (DNA)
phosphate group
nitrogen containing base
adenine - A
thymine - T
guanine - G
cytosine - C
uracil - U (RNA)
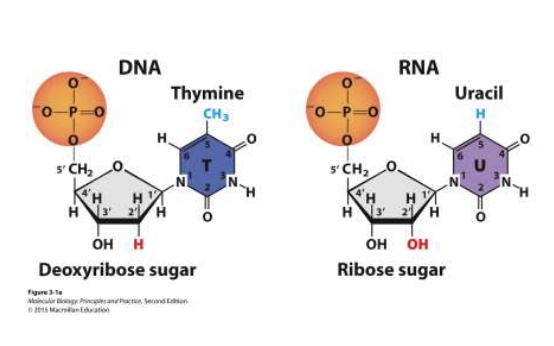
nucleoside
no phosphate (just sugar and base)
DNA/RNA bonds
phosphodiester bonds btwn 3’ OH from one n-tide and 5’ phosphate on next n-tide
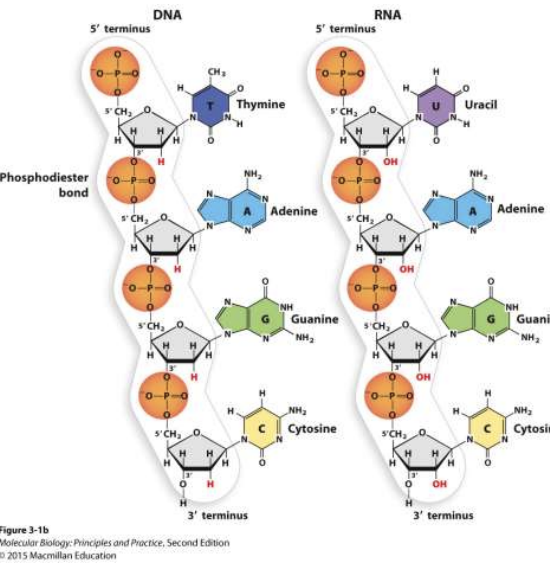
DNA vs RNA strands
DNA = double strand
RNA = single stranded
not always bcs complementary RNA strands can form double strand
backbone of nucleic acids
phosphate-sugar-phosphate-sugar, etc
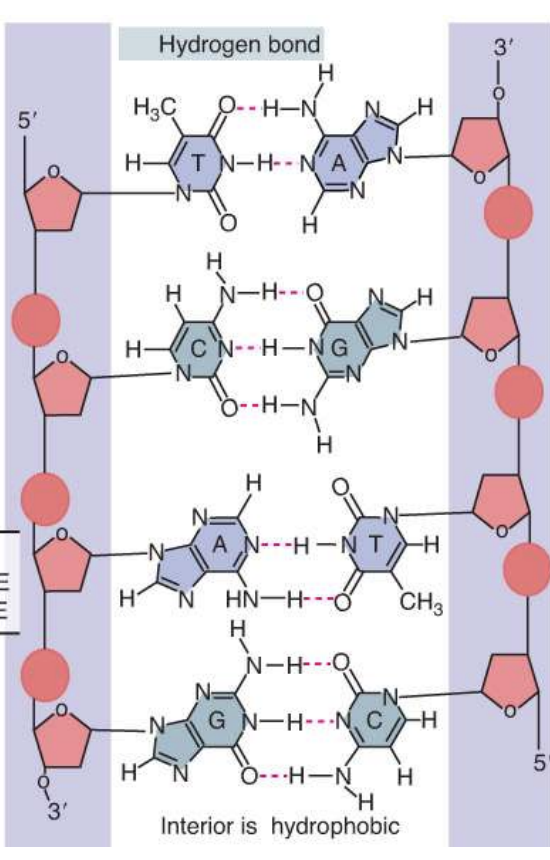
how can the number of base pair types (A-T vs C-G) on a DNA molecule be determined?
fewer H-bonds btwn A-T base pairs
will melt quicker as a result
can start by using lower annealing temps to see how much of DNA molecule disintegrates at a lower temp
grooves of DNA
major groove
where specific-base DNA binding proteins bind (because only place they can fit)
minor groove
only general DNA binding proteins ine because they cannot see the bases (since groove is too small) so they must be general
how many angstroms is each turn in the DNA double helix?
34 angstroms
what bonds hold the two DNA strands together?
hydrogen bonds
three forms of DNA
base pairs per turn of each
B-DNA: most common, right handed
10.5
A-DNA: favoured in solutions devoid of water, not sure if it exists in cells
11
Z-DNA: left handed, evidence that it is found in small stretches of DNA
12
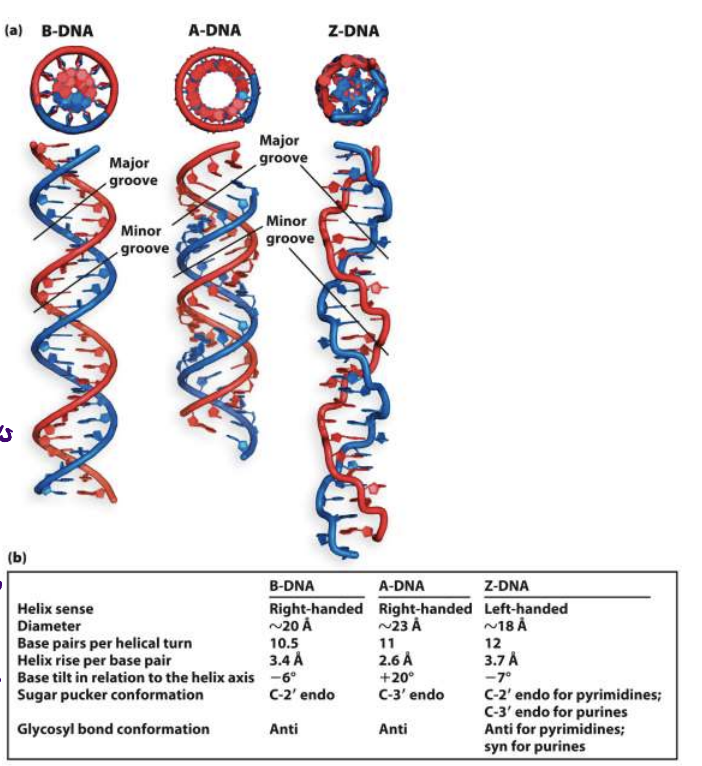
benefit to Z-DNA’s structure
grooves are bigger (even minor groove) therefore exposes bases better → easier protein binding
what is supercoiling and what does it do to DNA structure
supercoiling - coiling of closed duplex DNA in space so it crosses over its own axis (twists back on itself)
affects the structure
closed DNA is either circular DNA or linear DNA
ends are anchored so that they are not free to rotate
linking number
closed DNA molecule has a linking number (L), which is the sum of twist (t) and writhe (W)
L can be changed only by breaking and reforming bonds in the DNA backbone
twist and writhe
t - property of the double helical structure itself, representing the rotation of one strand about the other
represents total number of turns of duplex and is determined by number of base pairs per turn
w - in addition to this twisting, there can be extra twists or coils in the DNA called writhe
way to describe how much the DNA is "twisted" beyond its basic double helix structure
variation in DNA structure - backbone
rotation around contiguous bonds that make up sugar-phosphate backbone
nucleotides can either be linear (stacked on stop of each other) or puckered
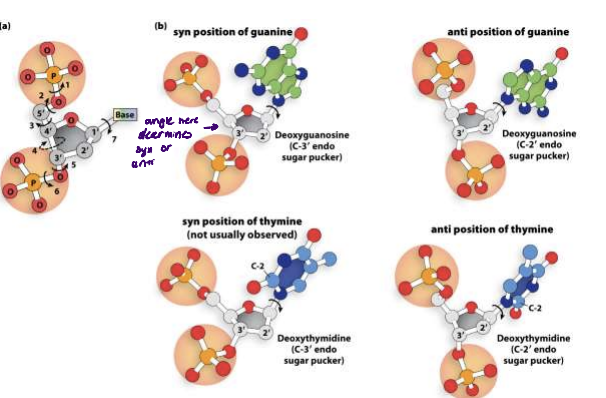
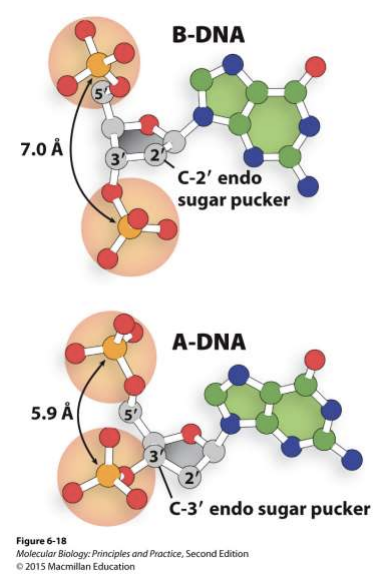
sugar pucker of DNA
A and B DNA differ bcs of sugar pucker
no water in btwn nucleotides in A-DNA allowing it to puker/sqeeze together
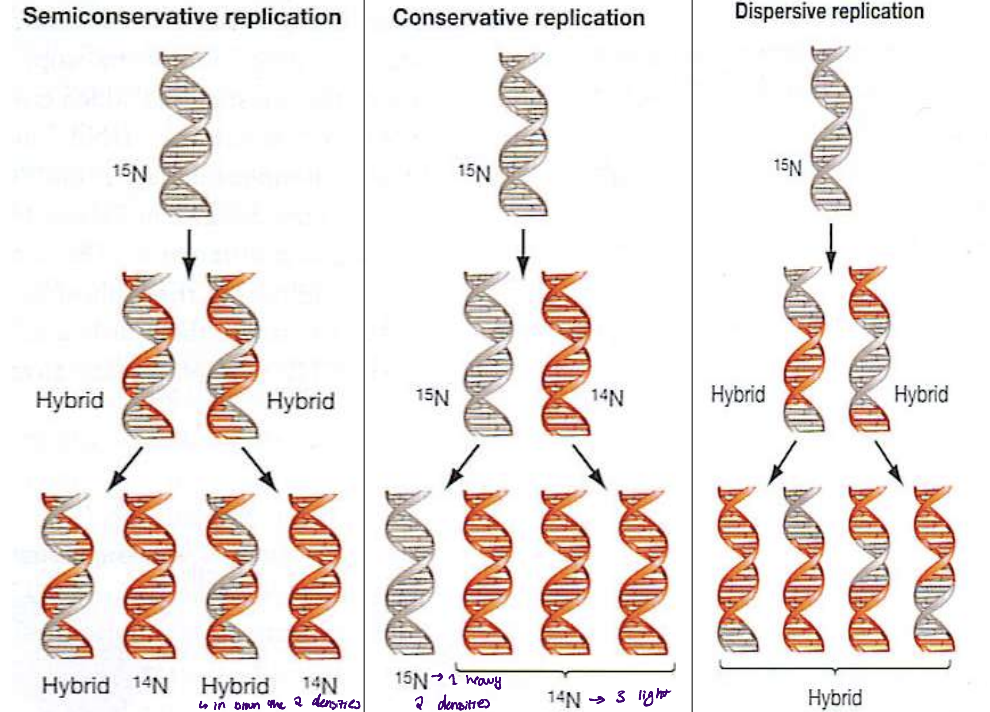
meselson-stahl experiment
purpose
what theories did they test
used radioactive N to label parental DNA molecules and proposed what they would see in each of the scenario
N15 - heavy
N14 - light
3 theories about DNA composition
conservative
semi-conservative
dispersive
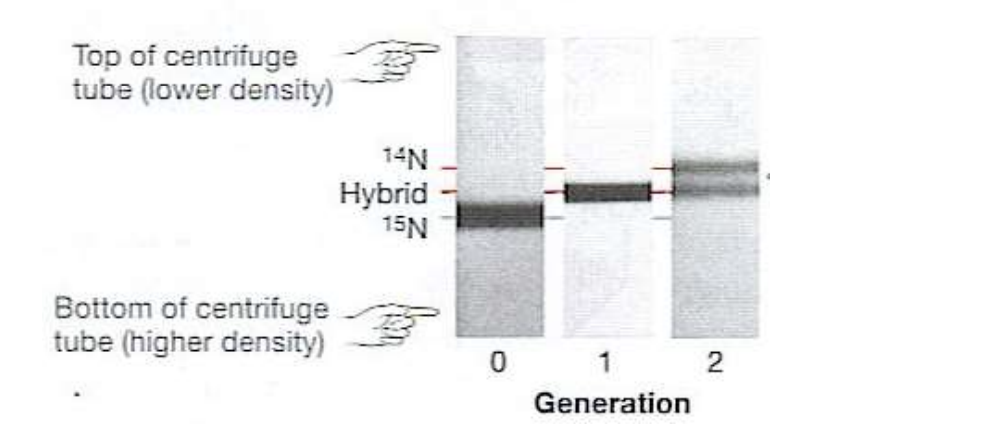
meselson-stahl experiment results
results show DNA exhibits
showed one hybrid, and one N14 band therefore its not conservative or dispersive, must be semiconservative
semiconservative replication
DNA replication accomplished by separation of the strands of a parental duplex, each strand then acting as a template for synthesis of a complementary strand
sequences of daughter strands determined by complementary base pairing w/ separated parental strands
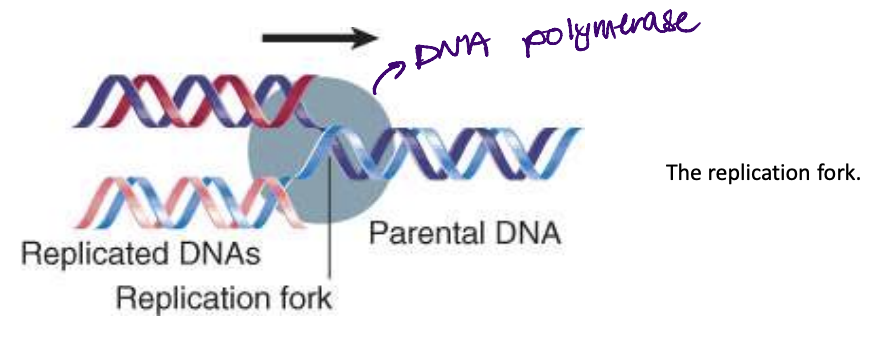
replication fork
replication fork- point at which the parental strands are separated
nucleases
enzymes that degrade nucleic acids; they include DNases and RNases
2 categories:
endonuclease - cleaves a bond within a nucleic acid (chews from within a strand)
example shows an enzyme that attacks one strand of a DNA duplex
exonuclease - removes bases one at a time by cleaving the last bond in a polynucleotide chain (chews from the ends)
can be 3’ or 5’ end

RNA polymerase
enzyme synthesizes RNA using a DNA template
formally known as DNA-dependent RNA polymerase
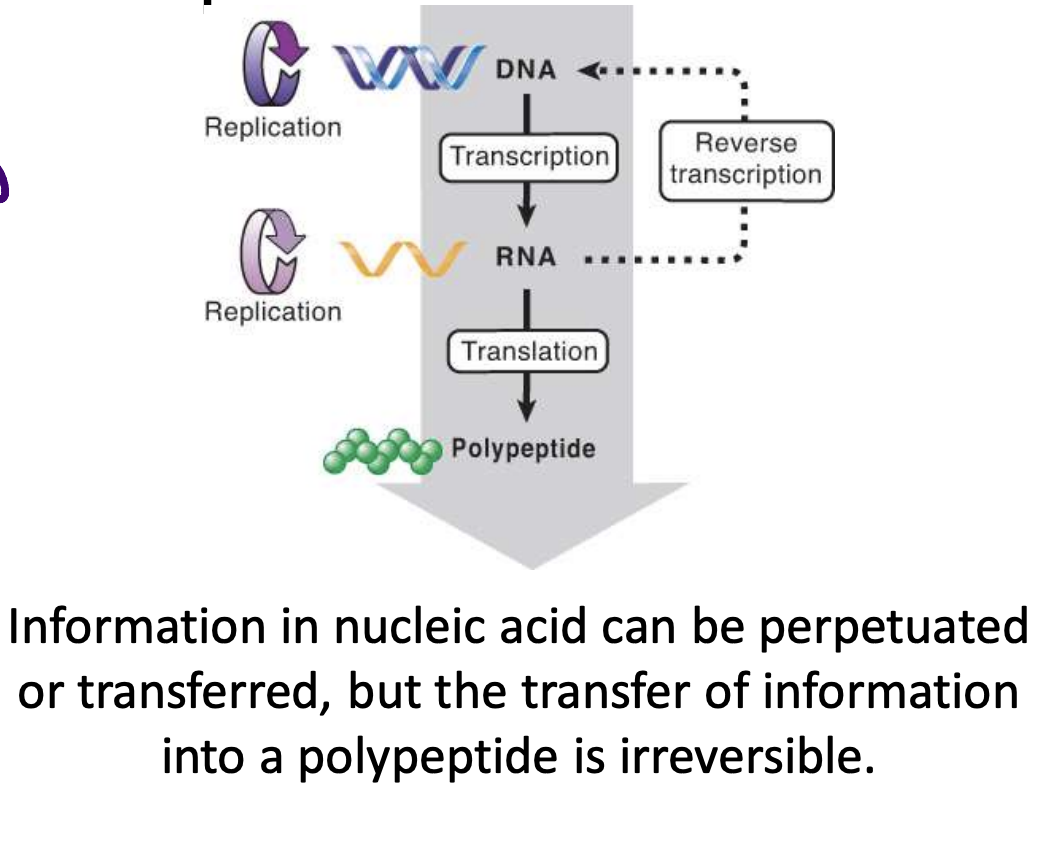
central dogma
info cannot be transferred from protein to protein or protein to nucleic acid, but can be transferred between nucleic acids and from nucleic acid to protein
translation of RNA into protein is unidirectional, can't be reversed (cannot go from protein to nucleic acid)
DNA can be unzipped and then re-zipped
explain
denaturation - involves the separation of the two strands due to breaking of hydrogen bonds between bases
renaturation - re-association of denatured complementary single strands of a DNA double helix
aka: annealing
happens when temperature is reduced after being risen
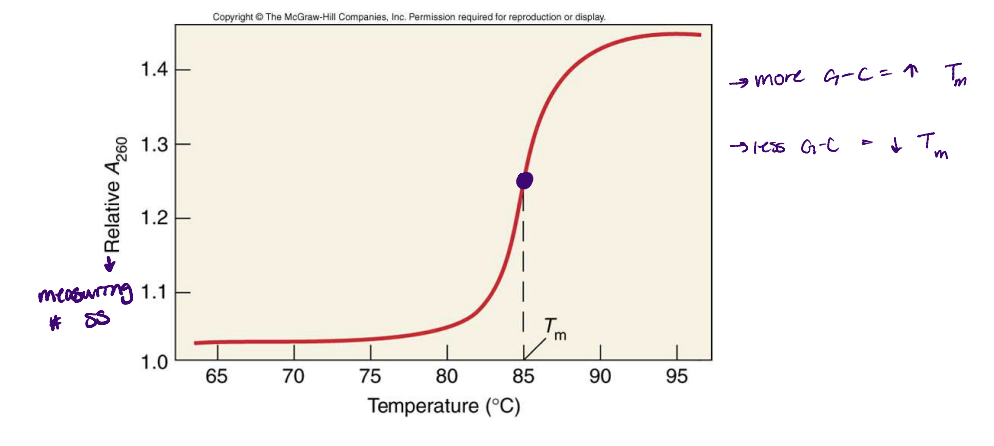
melting temperature (Tm)
is the midpoint of the temperature range for denaturation
50% of DNA has separated to 2 strands
depends on the G-C content → can be used to characterize genomes
nucleic acid denaturation and renaturation/hybridization
what molecules can be involved?
condition?
intramolecular or intermolecular?
DNA-DNA, DNA-RNA, or RNA-RNA combos
they just need have complementary base pairing
base pairing occurs in duplex DNA and also in intra- and intermolecular interactions in single-stranded RNA (or DNA)
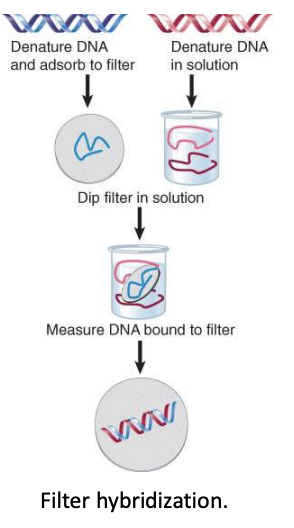
complementarity test - filter hybridization
ability of two single stranded nucleic acids to hybridize is a measure of their complementarity
can compare 2 genomes to look at overlap
c-value paradox
observation that more complex organisms will not always need more genes than simple organisms
most likely explanation for the paradox is that less complex organisms have more DNA that does not code for genes
more complex organisms means more complex gene regulation (because our genes can make multiple proteins)