Quant Genetics Final
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
88 Terms
what could a SNP in the exon cause
premature stop codon
frameshifts
missense stability
what would a SNP in the exon effect
mainly effect function of the gene product
protein stability
change of protein sequence
protein interaction
protein-dna interactions
what would mutation in the promoter region cause
promoter region is responsible for expression of the target gene
effects by changing a motif sequence, chromatin accessibility, transcription factor binding
how would you test for the causal effects of SNP in the promoter region
purify the normal and mutant proteins to test their functions in vitro
express protein in cell lines or model organisms to test function in vivo
how would you test for the causal effects of SNP in the exon region
transcription factor motif analysis to see if specific motif is disrupted by snp
luciferase assay to determine whether snp affect transcription
atac seq and histone modification chip seq to determine whether chromatin accessibility is affected or not
what are the 3 genetic explanations for the missing heritability problem
infinitesimal hypothesis: common variants have small additive effect sizes BUT responsible for most of var in the pop. (strongly supported)
rare allele hypothesis: rare alleles have large effects
epistasis hypothesis: extensive epistasis hidden in heritability studies (not well supported)
rare variants can have large effect on disease. give an example
GH1 deficiency
has large effect on height
but is rare → little effect on overall variance for height in human pop
we know that common variants have small additive effect but are responsible for most of the variance in a population + rare variants can have large effect but contribute little to variance.
explain this idea in terms of evolution
if mutation causes big harmful effect, natural selection will select against it (GH1 deficiency)
trait that is controlled by many variants (each with small effects), none individually is strong enough to trigger negative selection + preservation of genetic diversity
what is GCTA (Genome-wide Complex Trait Analysis)
using a mixed linear model
tool that combines the effects of ALL snps (not just the significant ones) to estimate h²
can severely overestimate or underestimate h² because population structure due to admixture
population admixture def
people from 2 dif ISOLATED places intermix
making a new mixed population
how do false positive occur from population admixture
failure to take mixed genetic background into account → false (+) in GWAS
why? alleles that distinguish the 2 pops will appear to be associated with the trait
what is protective varaint
genetic change that reduces the risk of developing a disease.
what is a haplotype
chunk of DNA variants (alleles or SNPs) where a set of variants are strongly linked
why? located close to each other on the same chromosome.
explain linkage disequilibrium and the problem that arises from LD
LD = physically close variants tend to be inherited together
why? closeness = less recombination
problem: some allele combinations are over represented → SNP might look associated with a trait but is really just “tagging along”
4 steps to find the responsible gene and mutation
extend association study to tracing inheritance in families
predict effect of mutation on gene func
stability? expression?
verify effect on gene func
Express normal and mutant proteins in cell lines and test functions.
Luciferase assays to test the promoter activity.
ATAC-seq and ChIP-seq for chromatin accessibility
verify effect on phenotype
gene knockout
what is cytological mapping and what do we use it for
technique to locate structural abnormality (deletions, inversions, translocations) on chromosome that may lead to
inactivate or inappropriately activate a gene
specifically we want to look at chomosoal abnormalities
when would cytological mapping come in handy
rare disease traits with not enough pedigree
Highly deleterious dominant traits
trait so harmful it does not get passed down aka the person dies
what are the two methods that we could use to help use identify mutations in the genome
cytological sequencing: sequencing of the entire genome
structural abnormalities (deletions, inversions, translocations)
exon seq: sequencing of just the exons
look for SNP in the protein coding region
what is exome sequencing and what do we use it for
much like cytological mapping we sequence only the exons and look for SNPs
use for single gene disorders that are
very rare disease traits with not enough pedigree
highly deleterious dominant traits (deadly)
what are the 2 things that could result in chromosomal abnomalities
inversions and deletions
translocation (result from breaks on different chromosomes)
what are some limitations of exon sequencing
relevant mutations are not necessarily on exons or protein coding regions
not useful for complex traits (trait that is influenced by many factors)
what is a SNP
a single nucleotide is changed
what are CPG islands and why do they matter
sequence of DNA with high CpG content
CPG islands are located in promoters
If CpG island becomes methylated by DNA
Methyltransferase, it usually silences the gene downstream
what is gene imprinting? why do we care? what are examples of diseases associated with imprinting?
1 gene is methylated (silent), 1 is unmethylated (expressed)
can either be the maternal or the paternal
Prader-Willi syndrome: loss of paternally expressed genes
Angelman syndrome: loss of maternally expressed genes
2 methods that control gene expression
histone modification
methylation
what does bisulfite seqeuncing do
tell you where the dna is methylated and not methylated
we talk about cis and trans acting regulation of gene expression. what does that mean
cis acting: promoters, enhancers, insulators
trans acting: transcription factors
what is a promoter
dna seq upstream of a gene
start site where RNA polymerase binds
essential for gene expression
think of it as an on and off switch transcription ex (TATA box)
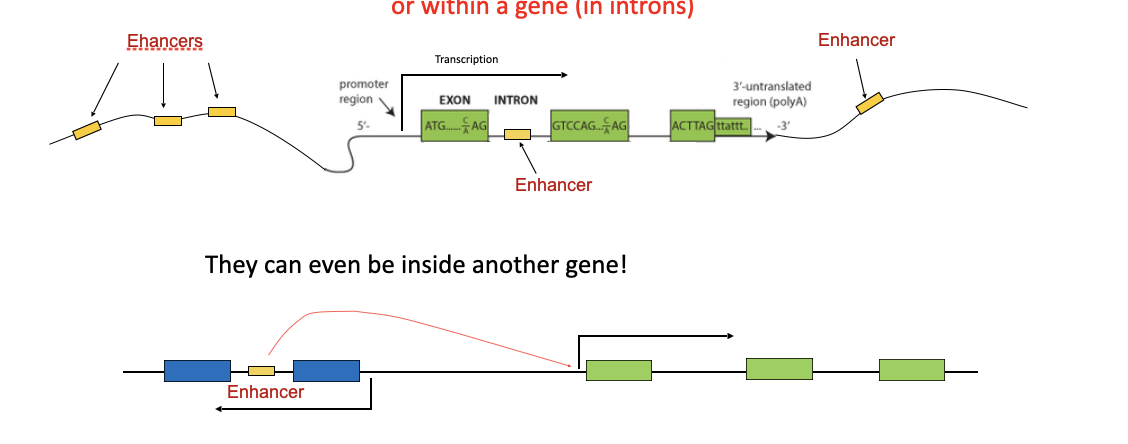
what is an enhancer
DNA regions that can boost the transcription of a gene
Enhancers bind transcription factors
Usually upstream of the promoter
but can be far upstream, downstream,
or within a gene (in introns)
A volume knob that amplifies gene expression — but only works when the right transcription factors are present.
what is a transcription factor
Proteins that bind to specific DNA sequences (in promoters or enhancers).
helps to activate or suppress transcription
what are locus control regions
An LCR is a long-range enhancer that controls multiple genes in a gene cluster.
think master regulator
what is chromatin
DNA wrapped around histone proteins
how do histones affect gene expression?
makes it hard for transcription factors and transcriptional machinery to access DNA…
what is epigenetics
study gene expression that do not involve changes to the DNA sequence itself ie methylation, Histone Modifications etc
chromatin states arise from DNA and chromatin modifications. what 2 things would result in open chromatin? condensed chromatin?
condensed: dna methylation, histone deacetylation
open: dna demethylation, histone acytylation
what does dna methylation mean? what is the function of DNA methylation? histone acetylation?
dna methylation: attaching a methyl group onto a cpg group (found in cpg island)
function: mechanism to control gene expression
histone acetylation: attaching an acetyl group, also control DNA methylation
what is the difference between de novo methylation and methylation maintenance
de novo methylation: establish a repressed state in response to developmental program or environmental factors
Maintenance methylation: maintains repressed state through somatic cell divisions
If methylation silences gene expression and it is heritable through cell division, how can new progeny develop?
Methylation status is controlled dynamically (erased during dev of germ cells then reset during subsequent differentiation of gametes, fertilization, and development.
what is the purpose of taking folic acid when women are pregnant?
folic acid increases methylation. deficiency in dna methylation is associated with neural tube defects.
when does imprinting occur
during germ cell differentiation
explain the parent of origin effects
disease phenotype is associated with the variant inherited from a specific parent
ex: UBE3A → angelman
explain angelman syndrome
UBE3A is methylated (silent) in father, but UBE3A gene is unmethylated (expressed) in mother
if mom’s is mutated/methylated there is NO UBE3A gene
result = angelman syndrome
what is interesting about prader-willi and angelman
mutation of the same chomosome 15
pws = mutation from father
as = mutation from mother
What is Uniparental Disomy (UPD)
two copies of a chromosome, or part of a chromosome, from one parent
and no copies from the other parent.
what are epigenetic mutation
disruptions in the regulation of epigenetic silencing
what are secondary epimutations v primary
secondary epimutation caused by real genetic mutation
DNA mutation
ex: mutation in a gene that regulates chromatin
primary epimutation that is Not caused by a DNA mutation
ex: improper methylation of a gene
what are 2 tools and databases for epigenetic
epigenomics roadmap project
encyclopedia of dna elements
what is functional genomics
study of how those dna seq behave in real biological systems
methylated dna is transcriptionally inactive/active
unmethylated dna is transcriptionally inactive/active
inactive
active
methods to identify region of methylated v unmethylated dna
bisulfite sequencing
Unmethylated cytosines → uracils
Methylated cytosines → stay as cytosines
tell me about bisulfite treatment
detection of methylated or unmethlated dna
extract dna
bisulfite treatment
PCR amplification & sequencing
C stays a C, it was methylated. C becomes a T, it was unmethylated
1 pro and con of bisulfite sequencing
detects methylation at both CpG and non-CpG sites.
reducing sequence complexity, which can make it difficult to create alignments
what is microarray hybridization and what does it do
comparing 2 dif samples to detect differences
2 dif samples of DNA or RNA are washed onto a hybridized chip. compare how strong the sample binds to the probe via dif colors.
method to detect regions of condensed chromatin v uncondensed chromatin
atac seq
DHSS
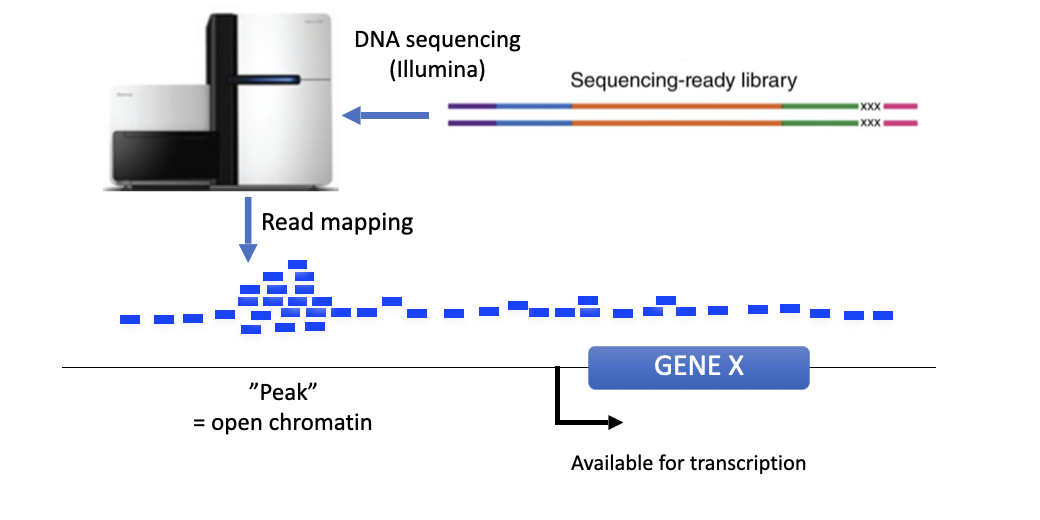
what is atac seq and explain
method for detecting open chromatin
pro: simple and fast sample prep
fresh tissue isolation
when reading the map of HSS and atac seq, what do the peaks indicate?
open chromatin
what is DHSS and explain
DNase Hypersensitive Site Mapping
dhs is site where dna is not wrapped around histone tightly
this means DNase I can access it and cut it
pro: great sensitivity at promoters
con: time consuming
methods to identify dna sequences at which specific proteins are bound
chip seq
cut and run
cut and tag
chip seq
method to find binding sites for specific TF
crosslink dna
digest
add antibody of interest
capture and wash the complex
undo cross link and seq the purified DNA
what would a peak from chip seq indicate
there is an occupied binding site
how could you use atac seq, hss, and chip seq together
atac seq and hss to identify areas of open chromatin
chip seq to determine sites that a protein binds to
they should both be the same region
methods for genome wide measurement of gene mrna expression
microarray
rna seq
what would you use rna seq for?
Use to compare gene activity
• in different cell types
• in disease vs healthy cells
• under different conditions
cancer is a heterogenous group of disorder. what does this mean
Each cancer type has its own causes, mutations, and behavior (breast cancer ≠ leukemia ≠ melanoma)
yet they share 2 common properties
uncontrolled cell proliferation (divide and grow)
cell spreading to nearby tissue
hyperplastic v dysplastic growth
hyperplastic: excessive number of cells that appear the same a those in normal tissue
dysplastic: growth that has abnormal cells
benign v malignant tumor
benign: no invasion, self limiting (absense of blood vessels limit size), can be removed surgery
malignant: tumor spread to neighboring cells, grow fast
what does the darwin model of tumor progression say
Tumors are not just masses of cells — they are dynamic, evolving populations of cells that accumulate mutations, compete for survival, and expand when advantageous mutations arise.
2 consequences
clonality tumors descend from a single ancestral cell
genetic heterogeneity = cells are dif and respond dif to treatment
2 types of cancer genetics
familial = hereditary = germline mutation
sporadic = not hereditary = somatic mutation (not germ cells)
characteristics of fam pedigree with sporadic cancer
few affected fam members
onset late in life
2 types of cancer related genes
proto-onco gene: promote cell proliferation and survival / dominant oncogenes
tumor supressor: prevent inappropriate cell proliferation or survival / recessive oncogenes
gain of function mutation v loss of function
gain: causes a gene product to do more than normal
loss: reduces or eliminates the normal activity
what type of mutation in proto onco genes would promote cancer
gain of function
3 ways that proto onco genes can be activated
mutation within the gene
multiple copies of the gene
gene moved to new dna locus = under new control
promoter fusions
activation of proto-oncogenes
occur because oncogene is placed next to a strong promoter or enhancer
ORF fusions
activation of proto-oncogenes
occur because orf of 1 gene is fused to another gene that is already highly expressed
what is interesting about cancer
recessive at the cellular level, (tested by the heterokaryon test)
but the inheritance pattern is dominant
2 hit hypothesis
heterozygous person (but norm func)
but only needs 1 more mutation event to lose Rb function completely (loss of heterozygosity)
then the cell gives rise to a tumor
dominant negative mutation
produces a mutant protein that actively interferes with the normal, wild-type protein.
reasons for dominance of oncogenetic loss of function mutations
dominant negative effects
haploinsuffiency
loss of heterozygocity
haploinsuffienciy of some tumor supressor genes
loss of 1 of the 2 functional alleles is not enough to maintain normal cell function.
what are dominant negative alleles
Dominant-negative alleles antagonize the function of the wild type allele
similar to the dominant negative effect
1 rotten apple ruins the bunch
heterokaryon test
test used to determine if a disease is caused by a dom or rec mutation.
fighting cancer the old way and it’s challenges
1 - surgery, radiation, chemo
2 - effectiveness (new cancer from therapy, incomplete removal), side effects (target only cancer)
target cancer drug
act on specific targets - likely proto oncogenes because inhibiting them should slow down growth of cancer
criteria for picking targets (using the targets treatment approach)
area with high expression of the protein
missense mutation (snp)
gene fusion
car t therapy for cancer
a person T cells are genetically modified to express a CAR
car = synthetic receptors that make the t cell recognize specific cancer markers
CAR T cells seek out and destroy cancer cells
ex: KYMRIAH
risk of car t cell therapy
neurological toxicity
cytokin release syndrome
limitation of car t cell therapy
highly specific antigens are not available for most cancers
sometimes attacks the healthy cells
work around: double gated car t: car t cell only becomes activated if 2 antigens on tumor cell match.