M7 Van- Containment: from lipids to membranes
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
11 Terms
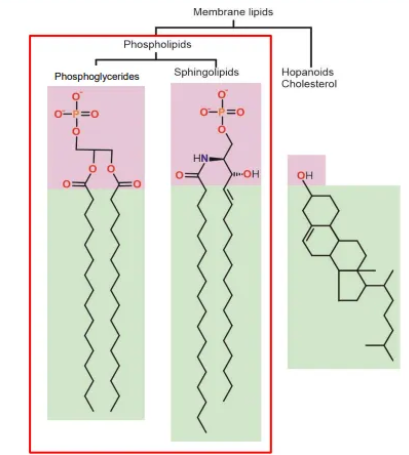
what are the three main types of membrane lipids?
phosphoglycerides (‘typical’ phospholipid)- glycerol linker, phosphate group + two fatty acids, all joined by ester bonds
sphingolipids (another phospholipid)- sphingosine linker, phosphate group, joined by an ester bond, + two fatty acids, joined by amide bonds
hopanoids + cholesterol- flat, hydrophobic molecules with saturated rings that intercalate into the bilayer and increase membrane stiffness
hopanoids are pentacyclic compounds, found in prokaryotic membranes
cholesterol is a tetracyclic compound, found in eukaryotic membranes
how can phospholipids vary?
variation in the tails:
tail length (longer = less fluid)
fatty acid saturation, normally only in one tail- cis double bonds are common, trans are rare (unsaturated = less tightly packed + more fluid)
variation in the heads:
head groups attached to the phosphate are involved in protein interactions, signalling and recognition
eg. glycerol, serine, glucose, choline, ethanolamine, inositol
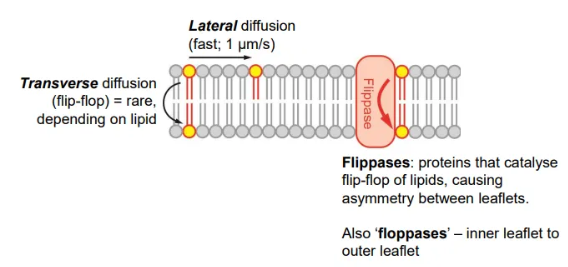
compare diffusion of lipids within and between membrane leaflets
diffusion within leaflets is very fast
diffusion between leaflets is rare, because it is difficult to get the hydrophilic head group past the hydrophobic tails, but can be catalysed by flippases
this causes asymmetry in the membrane, because there will be more phospholipids in one leaflet than the other
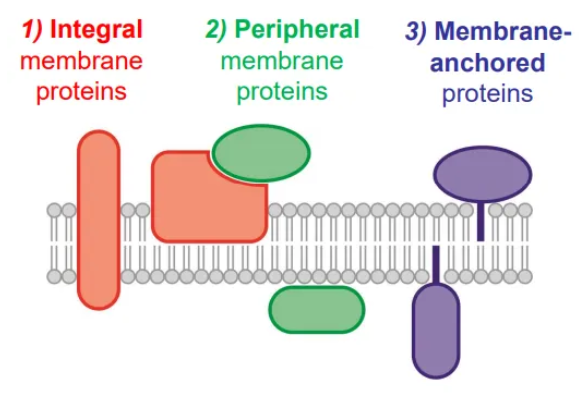
what are the three types of membrane protein?
integral membrane proteins have transmembrane domains (alpha helices, helical bundles and beta barrels) which have many hydrophobic residues that interact with the fatty acids
peripheral membrane proteins associate with membrane lipids and proteins via polar interactions, so they can be dirsputed by high salt concentrations
membrane-anchored proteins have lipid tails that are added post-translationally to interact with the fatty acids
in what three ways are membrane-anchored cytoplasmic proteins lipidated?
S-acylation- post-translational, reversible modification on cysteine residue by a thioester bond
N-myristoylation- post-translational or co-translational (during translation), irreversible modification of an N-terminal glycine residue (once methionine removed) by an amide bond
prenylation- post-translational, irreversible modification of cysteine residue near the C-terminus by a thioether bond
in what two ways are membrane-anchored extracellular proteins lipidated?
in prokaryotes:
lipoprotein- post-translational modification on N-terminal cysteine after a signal peptide has been removed
in eukaryotes:
GPI anchor- co-translational modification at C-terminus
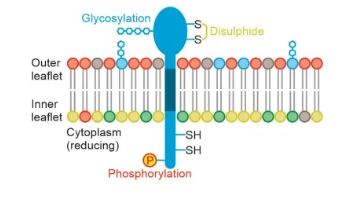
how and why are membranes asymmetrical?
different lipids aren’t evenly distributed between leaflets due to flippase action
proteins are asymmetrical and can’t flip
different PTMs are found on the cytoplasmic and extracellular sides of proteins (eg. outer environment is more oxidative, so disulphide bridges mostly form outside the cell)
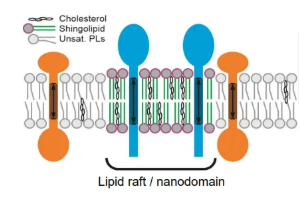
what are lipid nanodomains?
lipid nanodomains/rafts are localised membrane regions with distinct lipid compositions that can attract different proteins
these robust regions arrange membrane proteins into functional clusters, and alter local membrane rigidity
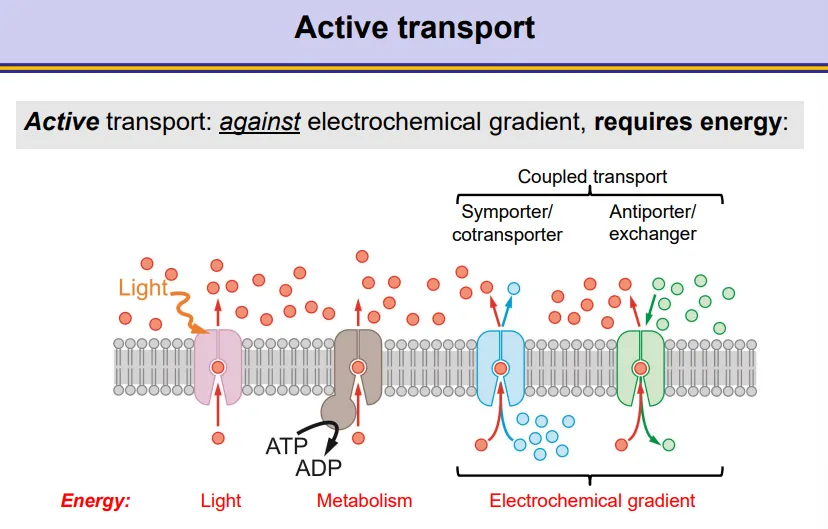
what are the different sources of energy for active transport membrane proteins?
light
ATP from metabolism
the electrochemical gradient of another molecule- coupled transport
symporters/cotransporters move the two molecules in the same direction
antiporters/exchangers move the two molecules in opposite directions across the membrane
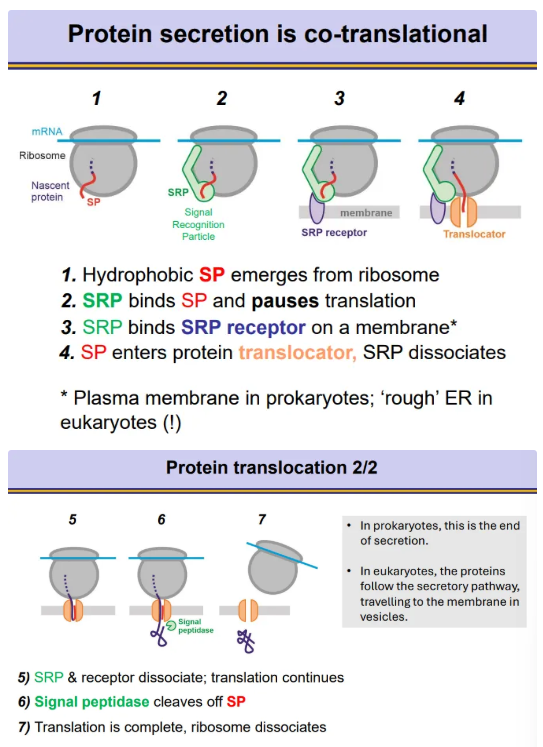
how are proteins secreted co-translationally?
proteins to be secreted contain a signal peptide sequence at the N-terminus
as the ribosome translates the protein, once the signal peptide emerges, a signal recognition particle (SRP) will bind, which pauses translation
the SRP binds to an SRP receptor on a membrane, so that the signal peptide can go through a translocator across the membrane
SRP dissociates so translation can continue
the signal peptidase enzyme cleaves the signal peptide off at the end
in prokaryotes, the membrane is the plasma membrane, so the protein is excreted straight out the cell
in eukaryotes, the membrane is the rough ER, so the proteins must go through the secretory pathway for excretion via vesicles
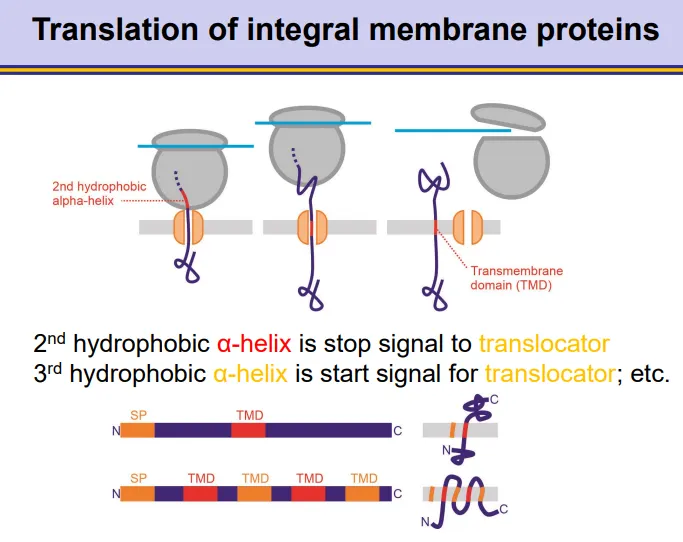
how are membrane proteins integrated co-translationally?
integral membrane proteins contain a signal peptide sequence at the N-terminus
as the ribosome translates the protein, once the signal peptide emerges, a signal recognition particle (SRP) will bind, which pauses translation
the SRP binds to an SRP receptor on the plasma membrane, so that the signal peptide can begin to go through a translocator across the membrane
SRP dissociates so translation can continue
when hydrophobic alpha helices are translated, these will get recognised as a transmembrane domain
the signal peptidase enzyme cleaves the signal peptide off at the end