molecular bio
1/242
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
243 Terms
genome
all genetic info, almost every cell in body contains entire genome
genes hold code to create proteins
transcriptome
all RNA
may vary over time
made up of
mRNA (1-2%)
ribosomal RNA (80%)
tRNA (15%)
micro RNA
small nuclear RNA
proteome
the protein
each cell has varied requirements so proteins expressed can vary
flow og genetic info
DNA → RNA → Protein
DNA replication
DNA → DNA
transcription
DNA → RNA
translation
DNA / RNA → Protein
types of proteins
proteins fold into various structures
ion channels
enzymes
trancription factors
receptors
antibodies
nucleic acid purines
adenine, guanine
have 2 rings
nucleic acid pyrimidines
cytosine
thymine (DNA)
uracil (RNA)
have 1 ring
chargaff’s rules
purines bind to pyrimidines
base pairing (at physological pH)
A - T (2 H bonds)
G - C (3 H bonds)
base pairing (at low pH)
A - T ( 1 H Bond)
G - C (2 H bonds)
extra protons acidify some areas, preventing binding between bases
sugar component (RNA, DNA)
1’ = N-glycosidic bone attaches
2’ - OH for ribose, H for deoxyribose
3’ = hydroxyl (OH)
5’ = phosphate attachment site

sugar-phosphate backbone
overly negative due to phosphates
phosphodiester bond - between the sugar and phosphate
N-glycosidic bond - between the sugar and base
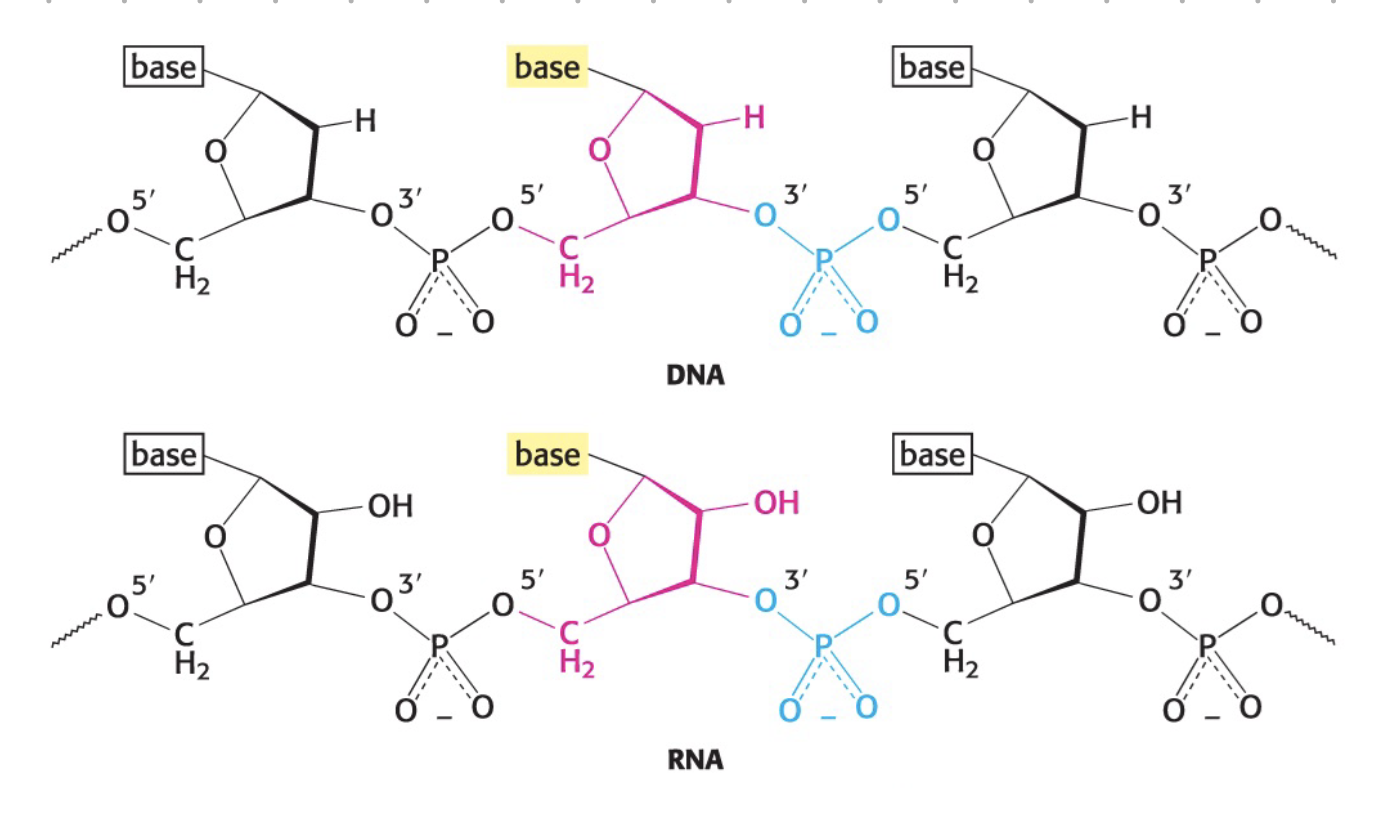
backbone structure
base attaches to 1’ of sugar = N-glycosidic bond
phosphate attaches to 4’ and 3’ of sugar = phosphodiester bond?
phosphate end = 5’
hydroxy end = 3’
interactions forming DNA helix
H bonds
ionic interactions
van der waals
hydrophobic interactions
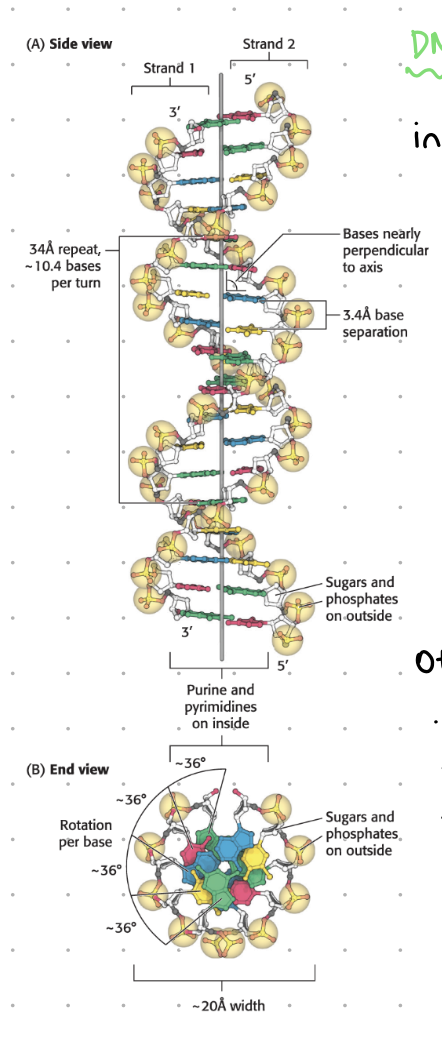
H bonds - DNA helix
between complimentary bases
2 for A-T
3 for G-C
ionic interactions - DNA helix
backbone has many -ve phosphates, repel to max distance creating helix twist structure
hydrophobic interactions - DNA helix
pyrimidines and purines are central, while hydrophobic sugars and phosphates remain on outer side
base stacking/vaan der walls - DNA relix
van der waals favours base stacking
the distance between bases is 3.4 A (radius of C = 1.7A) leading to interactions
helix properties
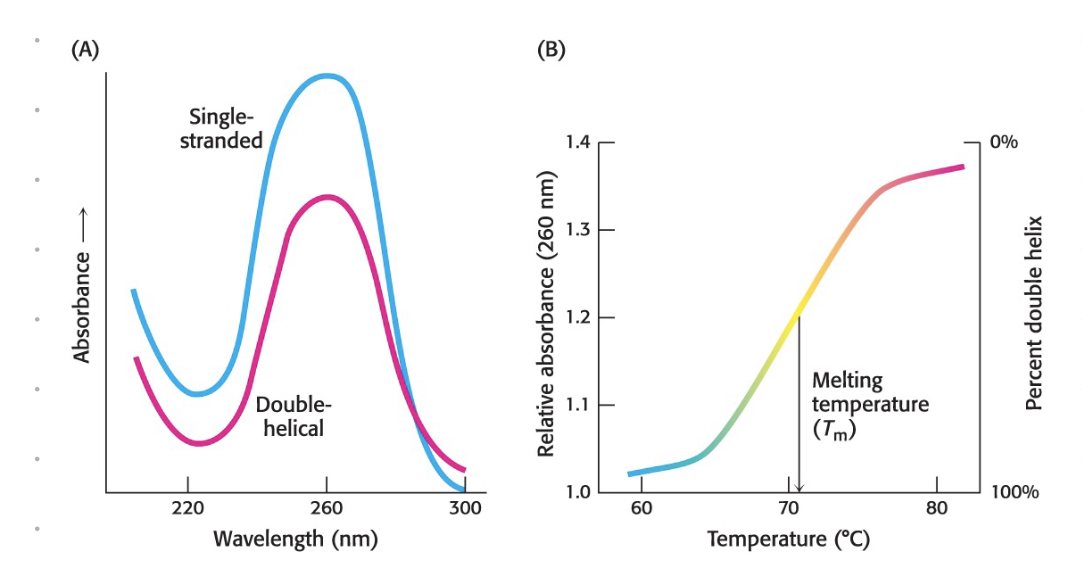
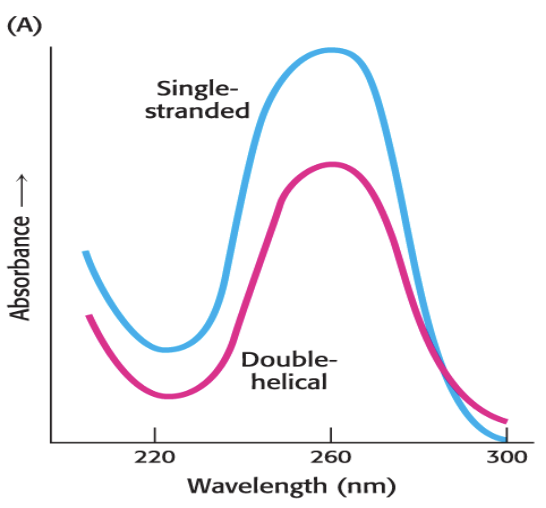
all bases absorb at 260 nm
[DNA] of 50 ng/mL = absorbance of 1
[RNA] of 4 ng/mL = absorbance of 1
aromatic protein rings absorb at 280 nm
ratios used to determine purity + contamination
hyperchronic effect
due to base stacking
double stranded DNA absorbs less at 260 nm compared to RNA
major + minor grooves
due to the way the bases bind to the backbone
base pairs further for major groove
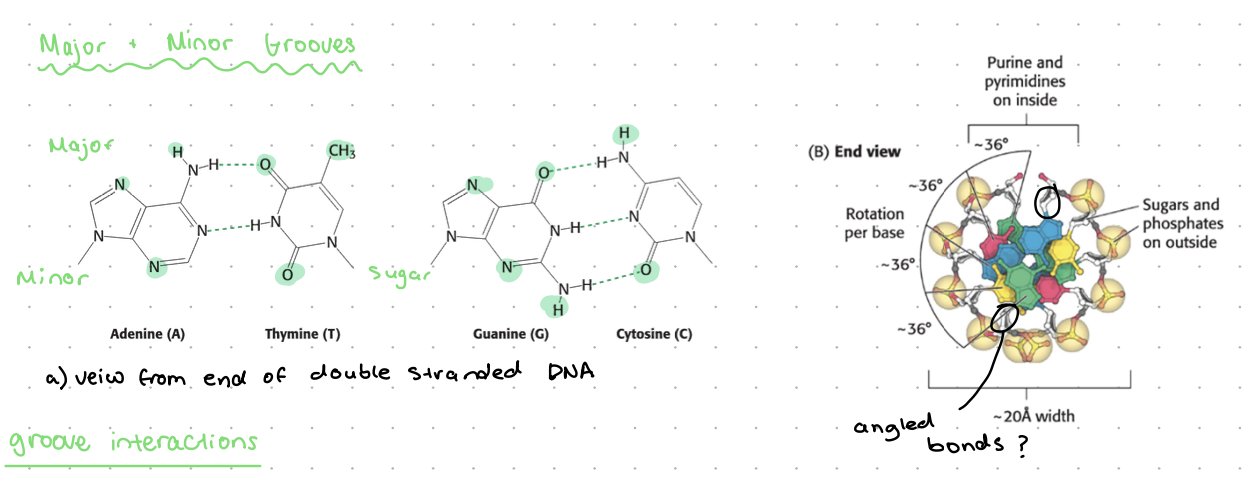
DNA groove interactions
binding of zinc finger transcription factors (or other proteins) to major groove
smaller things such as DAPI (DNA dye) can bind to minor groove
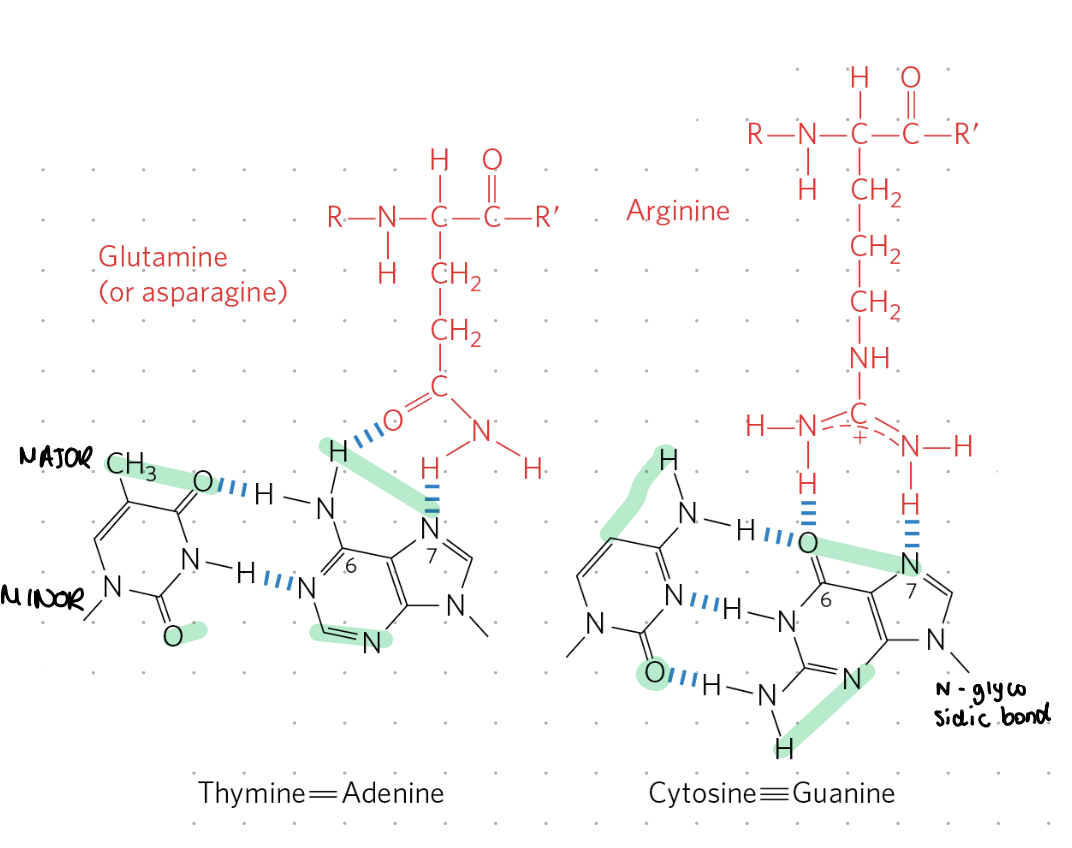
storing genetic info
hydrophobic bases contain info, protected inside helix
DNA is double stranded so a backup code is accessible
DNA vs. RNA
2’ of sugar, H for DNA, OH for RNA
uracil = RNA, thymine = DNA
RNA in acidic pH may degrade
RNA stability varies between cells
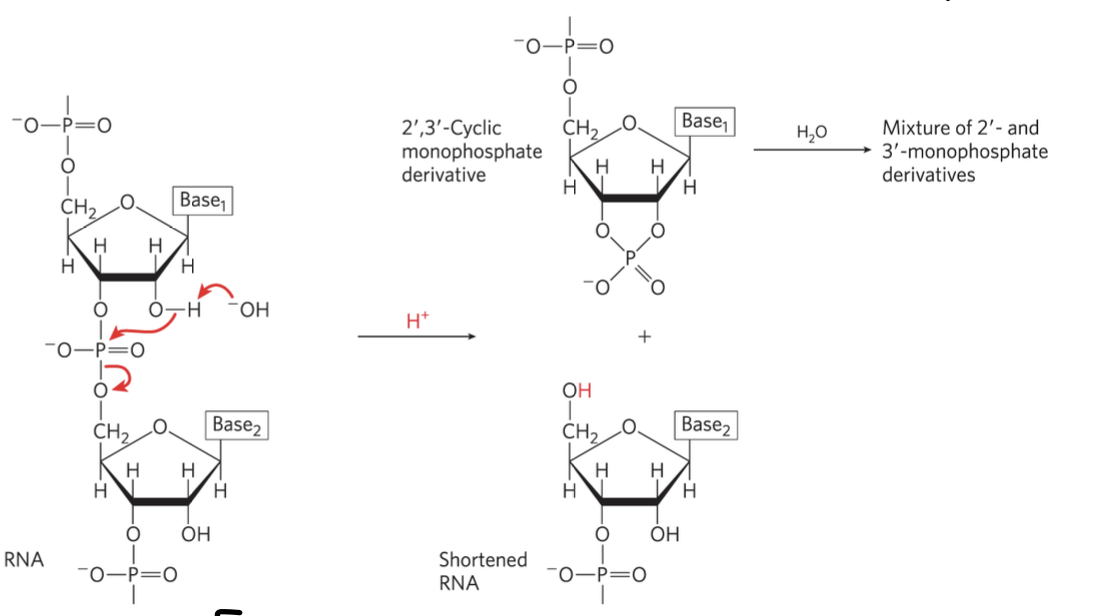
uracil in DNA - problem
cytosine may spontaneously deaminate into uracil
this can be recognised and removed in DNA
this issue is not corrected in RNA, but may lead to mutations
occurs 100-500 times per cell per day
E. coli
grows very quickly, very little needs
used often in molecular bio
one circular chromosome of 4.6 million bp
must copy entire genome to replicate (replication), occurs in less than 40 mins
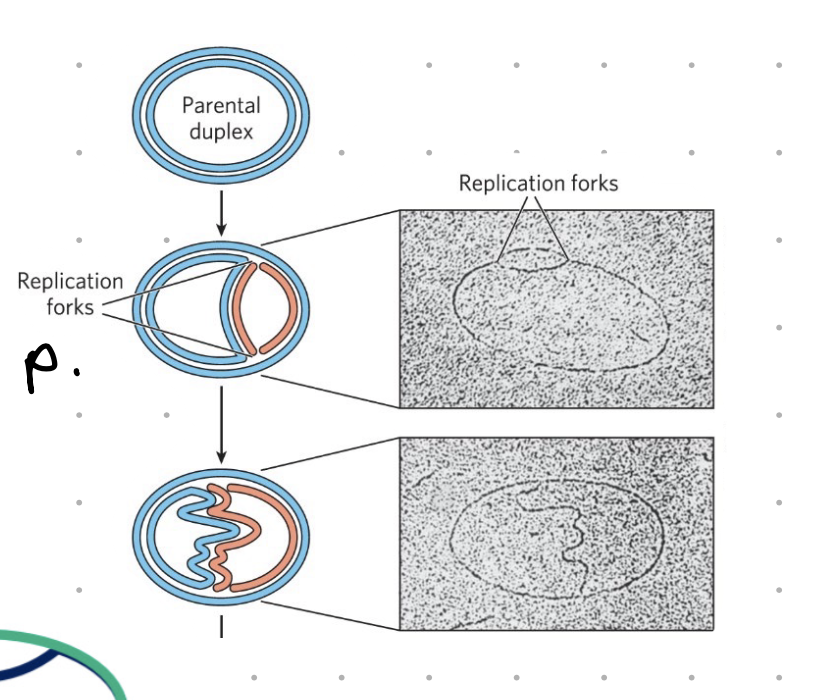
DNA → DNA (briefly)
separate DNA strands
primers bind to complimentary sequence
complementary base H bonds
nucleotide added to strand
optional = proofreading
DNA replication rules
nucleotides added in 3’OH to 5’P direction
DNA polymerase adds nucleotides to the strand
result = 2 double stranded DNA molecules
each contains 1 OG strand, 1 new strand
“semi conservative replication”
copying DNA in E. coli
separate DNA strands (starting at ORI site)
primers bind to complementary sequences (primer = short RNA strand)
complementary base H-bonds
nucleotide added
proofreading
continues along strand until complete
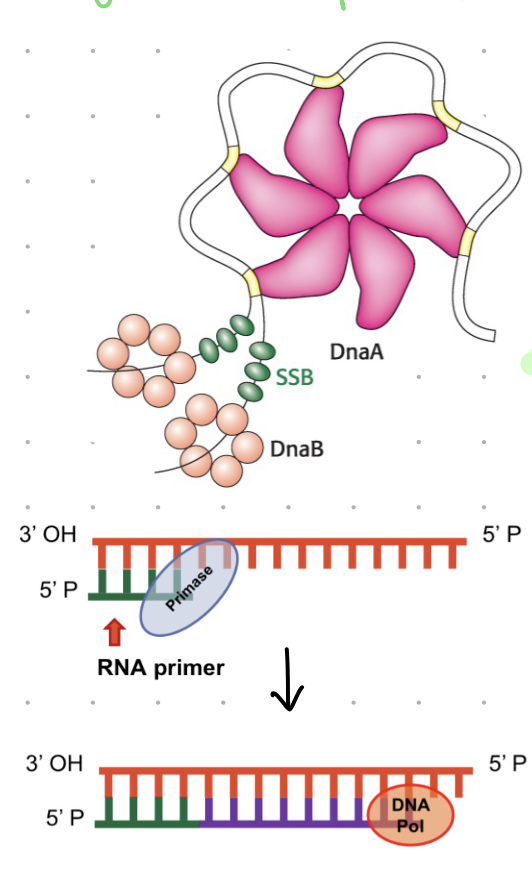
DnaA
bacterial protein that recognises oriC site (origin of replication)
acts as replication initiator by binding to a specific sequence (ORI) and initiating unwinding of DNA
prokaryotic replication
helice (aka DnaB)
moves 5’ to 3’ unwinding DNA
SSBP
single stranded binding proteins
keep DNA strands separated
primase (DnaG)
an RNA polymerase that makes short RNA primers
all DNA polymerases need a primer to add the nucleotides to
primase provides the 3’OH that nucleotides will bind to
note:
RNA polymerase do not need primers for synthesis
DNA polymerase used RNA primer for DNA synthesis
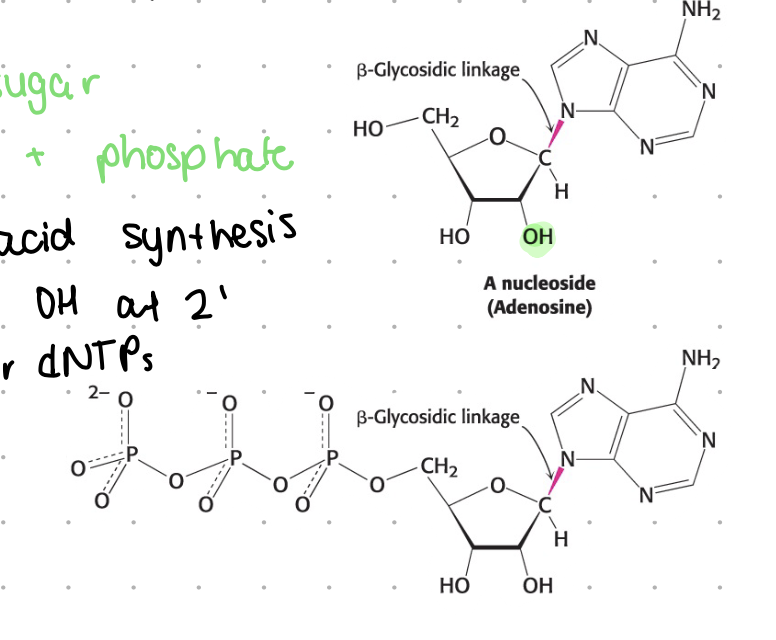
nucleoside
base + sugar
substrate for nucleic acid synthesis
for DNA, H instead of OH at 2”
generalize to NTP or dNTP
nucleotide
nucleoside (base + sugar) + phosphate
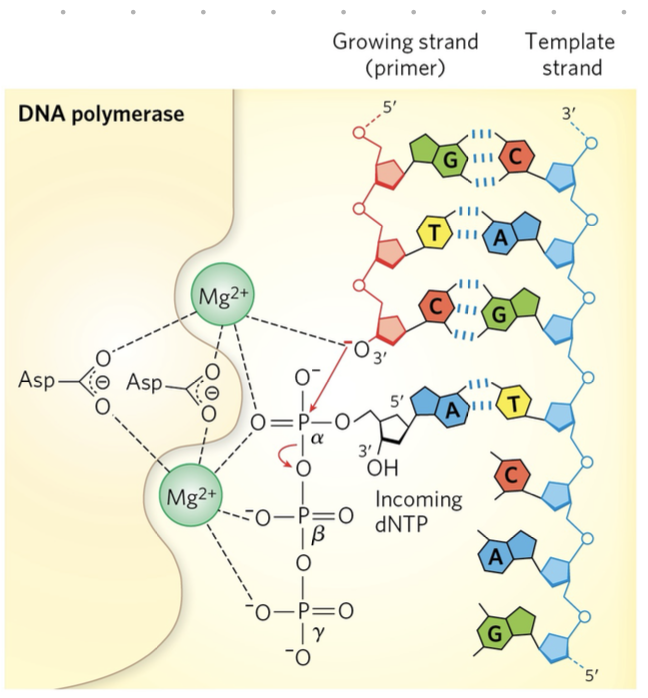
forming a phosphodiester bond (DNA)
base pairing w/ template strand
DNA polymerase catalyses formation of bond
pyrophosphate released
pyrophosphate further breaks down into 2 phosphates
released phosphates provide energy for next reaction (coupled)
base pairing must be…. during replication
checked/proofread
must be A-T/U or C-G
RNA/DNA syntheisis if thermodynamically unfavourable so
free energy released during the formation of phosphodiester bond provide the energy for DNA synthesis
forming a phosphodiester bond (RNA)
same as in DNA replication, although RNA polumerase used to catalyse bond formation rather than DNA polymerase
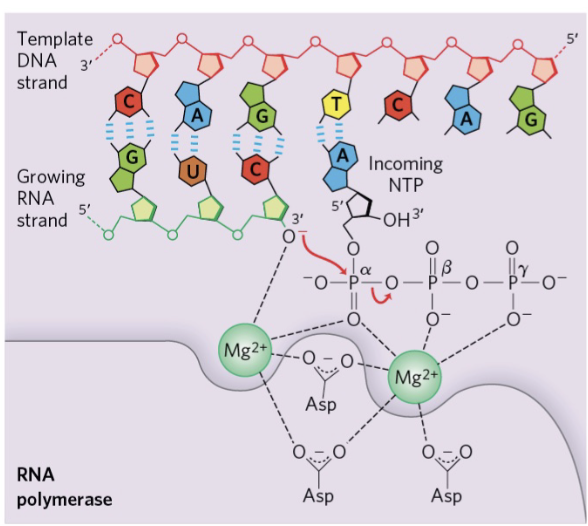
proofreading of base pairing
double check what base was just added, was it the correct
if not - can cut of by 3’ to 5’ (opposite) exonuclease
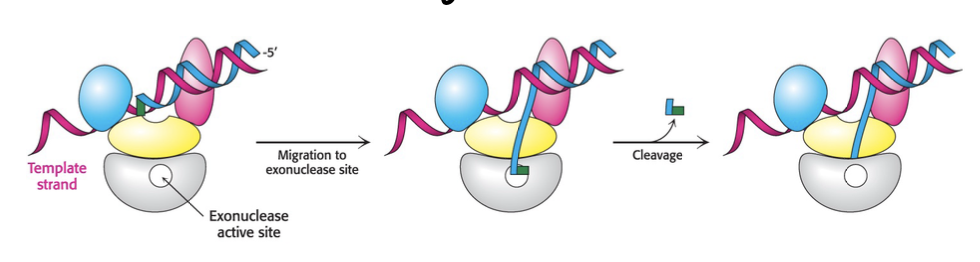
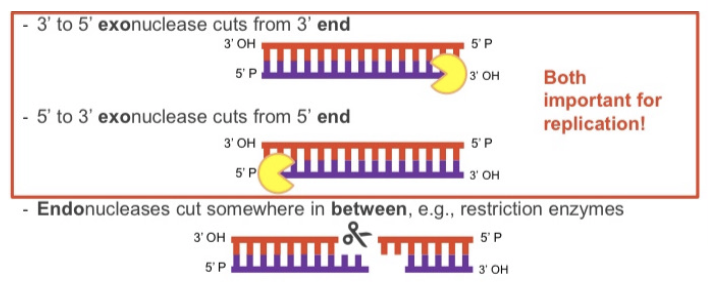
nucleases
enzymes which cut phosphodiester bond
3 to 5’ exonuclease cuts from 3’ end
5’ to 3’ exonuclease cuts from 5’ end
endonucleases cut somewhere in between
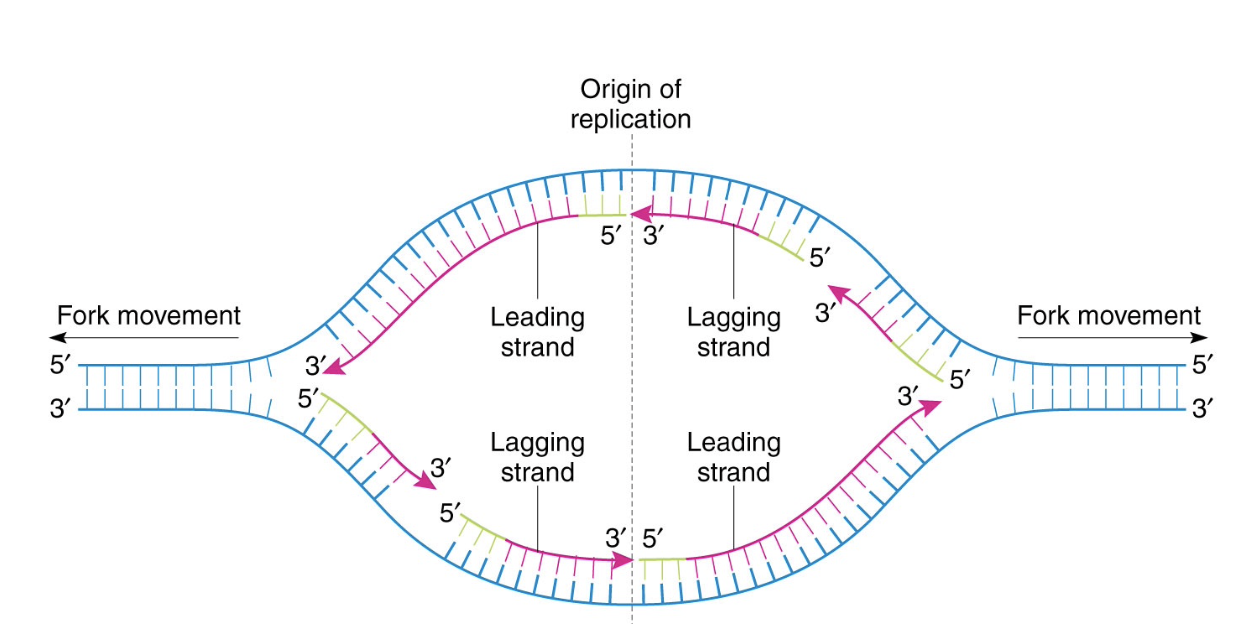
replication fork
A replication fork is a structure that forms during DNA replication, where the double-stranded DNA molecule is unwound and separated into two single strand
as helicase moves along each strand, the replicationbubble grows
DNA synthesised on both strands as each replication fork as it opens
issue - synthesising in the other direction
the lagging strand
synthesized as discountinuous okazaki fragments
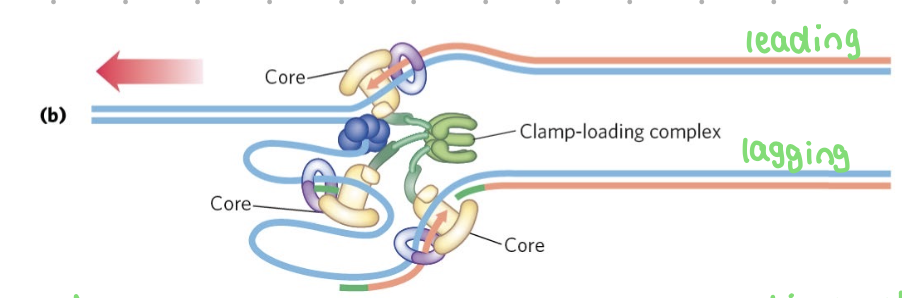
DNA pol III
core - adds nucleotides + proofreads
beta sliding clamp - holds tightly onto DNA - synthesis stops when it disassociates
clamp loader - grabs lagging strand + moves it to the polymerase
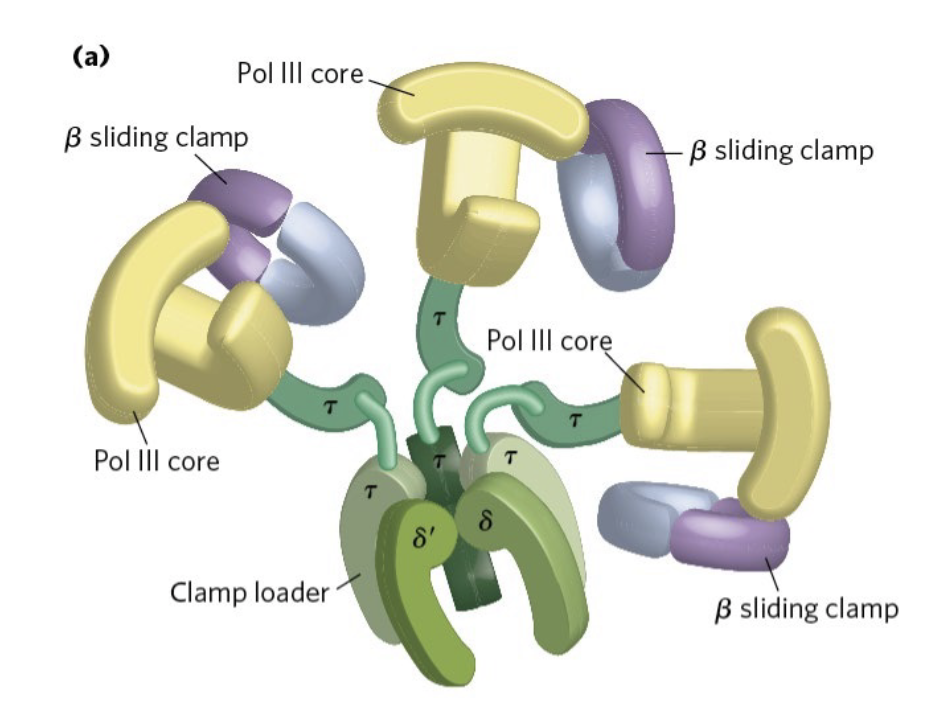
challenge - supercoiling
as DNA strands separate, it over winds = positive supercoiling
solutions = DNA topoisomerase 3
positive supercoling
twisting in the same direction as double helix, becomes tighter + harder to spearate
negative supercoiling
twist in opposite direction to the double helix - makes DNA easier to separate
topoisomerase 3
moves ahead of rep bubble + introduces negative supercoils
twists in opposite direction
cuts both strands, reorients ends + sticks them back together
requires ATP
removing primers
at end of replication, primer must be removed and replaced with DNA
done by DNA pol 1
DNA pol 1
removes RNA primer
has proofreading ability
has 3’ to 5’ exonuclease and 5’ to 3’ exonuclease abilities
can degrade RNA and incorporate DNA

DNA ligase
forms bond in backbone after primer removes, connects strands
termination of replication (prokaryotic)
it is important entire chromosome only copied once
mechanisms (TerA to TerK) prevent further synthesis
the 10 Ter sequences bind to a protein (Tus), creating trap for the fork
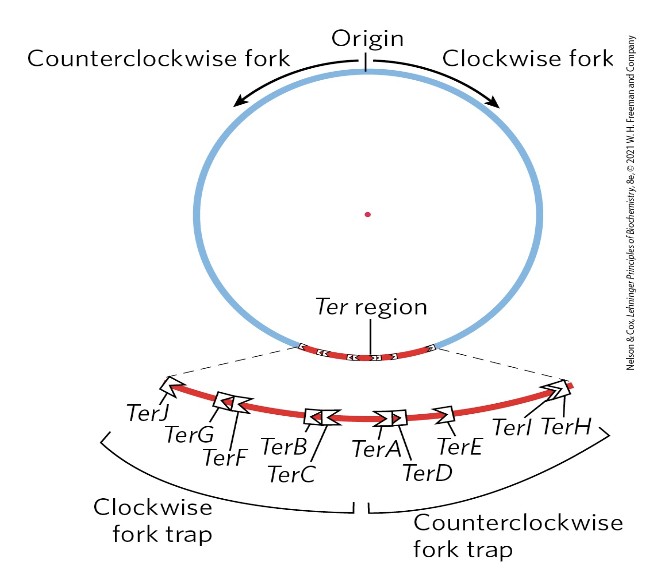
chromosome separation
another topoisomerase plays a role (type IV)

acyclovir - antiviral
treats herpes simplex virus infections
nucleoside analogue (modified nucleoside)
viral thymidine kinsae phosphorylates the drug to form nucleotide incorporate into growing strand in infected cells
azidothymidine - antiviral
HIV/AIDS streatment
nucleoside analoguse
viral reverse transciptase will incorporate drug into growing strand
molnupiravir
COVID-19 treatment
ribonucleoside analogue
once phosphorylated, viral RNA dependent RNA polymerase incorporates into the growing strand and keeps going
rationale - introduces mutations to point it can no longer work
5-fluorouracil (adrucil) - cancer drug
sugar + phosphate added in cell
thymidine synthetase methylates C-5 to convert dUMP to dTMP
when F5dUMP gets stuct, no dTMP is made
run out of substrate (dTTP) for DNA synthesis
only affects rapidly dividng cells
eukaryotic vs. prokaryotic replication (DNA)
mostly similar
initiation regulated - entire genome must be copied during replication
bi-directional
primase lays down RNA primers
highly processive polymerases do most of synthesis (3’ to 5’ exonucleoase activity)
leading + lagging strands
differences
1 circuler chromosome (pro)
many linear chromosomes (eu)
human genome
~3 billion x 2 bp
23 linear chromosomes
e. coli genome
4.6 mill bp
1 circular chromosome
controlled cell division
in eukaryotes
processes only occur if there are signals telling things what to do
e. coli
can divide quickly if provided with correct media + necessary nutrients
the cell cycle
the life cycle of a cell, encompassing its growth, DNA replication, and division into two daughter cells.
It's a coordinated process that ensures the accurate distribution of genetic material and division of the cell. The cell cycle is broadly divided into two phases: interphase and the mitotic (M) phase.
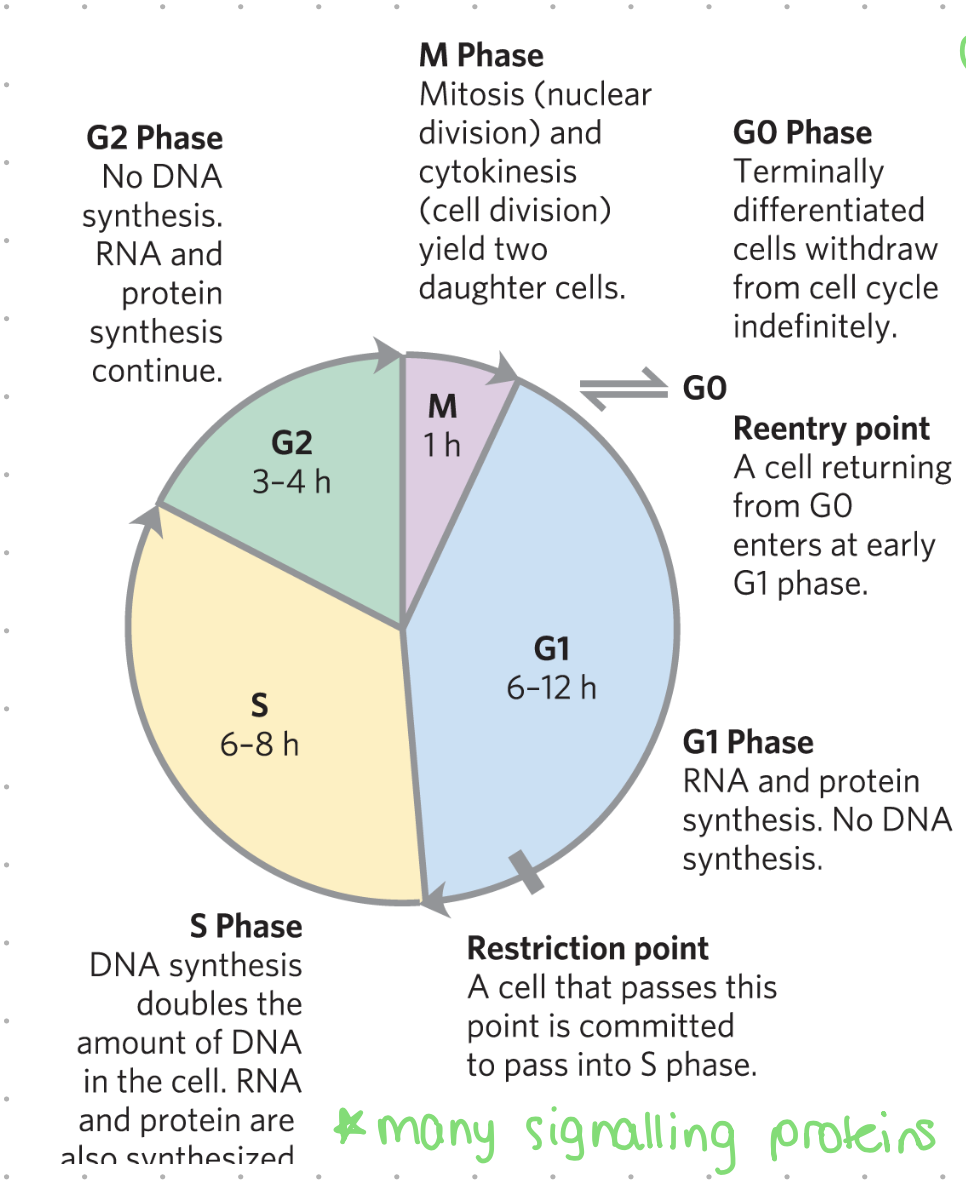
G1 (gap1) - the cell cycle
cells decide whether to divide (enter phase S) or stop (enter phase G0)
RNA + protein synthesis, no DNA synthesis
may last 6 - 12 hrs
G0 phase - cell cycle
cells not dividing
% of cells in G0 increases w/ age, also differs between cell type
terminally differentiated cells withdraw from cell cycle indefinitely
fibroblasts + epithelium
almost never enter G0 phase
adult liver cells
enter cell cycle ~ 1/ year
adult brain cells
almost always in G0 phase
quiescent cells
can be induced to re-enter cycle by mitotic signals
senescent cells
cannot re-enter cell cycle
restriction point
a cell that passes this point is committed to pass into S phase
S (synthesis) phase - cell cycle
DNA synthesis - doubling amount of DNA in cell
RNA and proteins also synthesized
many signalling proteins involved
may last 6-8 hrs
G2 phase - cell cycle
no DNA synthesis
RNA and protein synthesis continue
preparation for M phase (mitosis)
may last 3-4 hrs
M (mitosis) phase - cell cycle
mitosis (nuclear division) and cytokinesis (cell division) yielding two daughet cells
final phase of cell cycle
CDKs + cyclin
activation of kinases
levels may vary at phases
active CDKs phosphorylate proteins involved in regulation of cycle
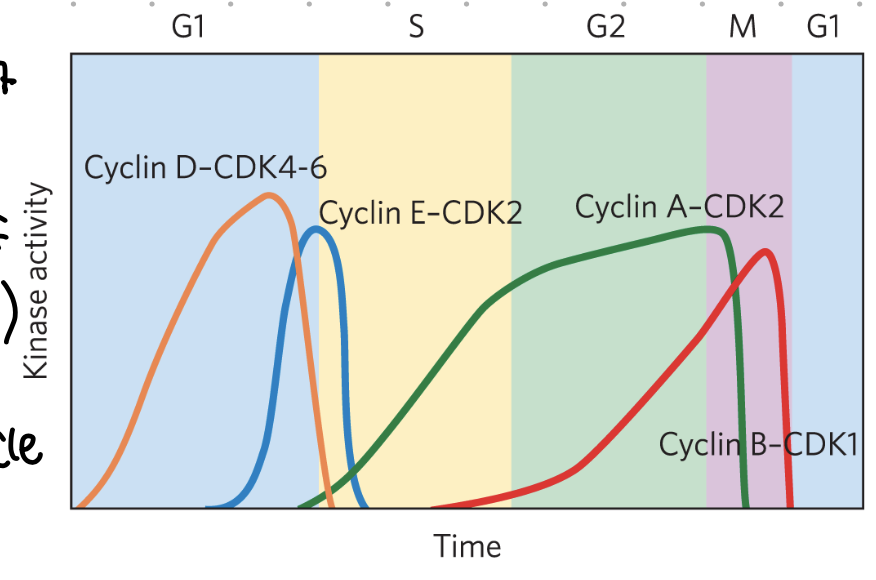
cell cycle checkpoints
G1 to S - check for DNA damage
S to G2 - check for DNA damage, and that synthesis is complete (no okazaki fragments etc)
M - check sister chromatids are correctly attached to spindles before division
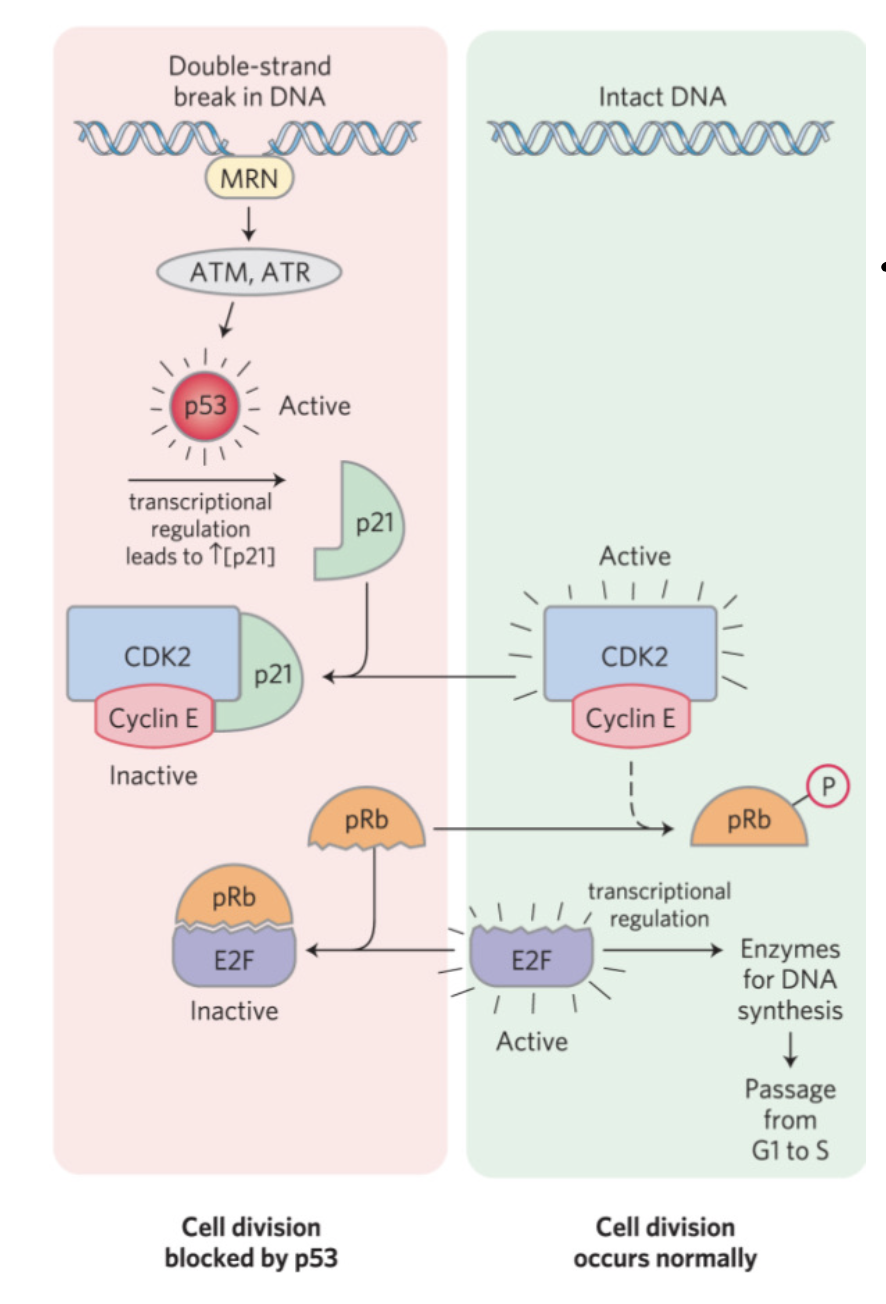
cell cycle checkpoint 1 (G1 to S)
important to control progression through the cycle
DNA must not be synthesized if damaged
DNA must be complete before progression to G2
check spindles before division
phosphorylation of target proteins by active CDKs affect function
oncogenes
“accelerator” proto-oncogenes control cell growth
referred to oncogenes when mutated
mutations are dominant, one copy is enough
Oncogenes are mutated genes that can lead to cancer. They are derived from normal genes called proto-oncogenes, which play a role in cell growth and division. When a proto-oncogene becomes an oncogene, it can cause uncontrolled cell growth and division, potentially leading to tumor formation
tumour suppressor genes
“the breaks”
slows down division or induces apoptosis
mutations are recessive - problems occur when 2 alleles
cancer
DNA mutations leading to cancer may be inherited (Rb, BRCA1) or acquired (behaviours - sunbathing, smoking, radiation, random during cell division)
retinoblastoma protein
a tumour suppressor protein
retinoblastoma = retina cancer in children
those w/ condition have often inherited one mutated copy and acquire a mutation in the other copy in retinal cells → two copies
people with mutaiton also have increased risk of lung, prostate and breast cancer
eukaryotic replication unique differences
much more genetic material to copy
much slower
many ORI sites - not all are activated in each round of replication, but enough need to be activated to copy all chromosomes entirely
regulated to ensure rep only once per cell cycle
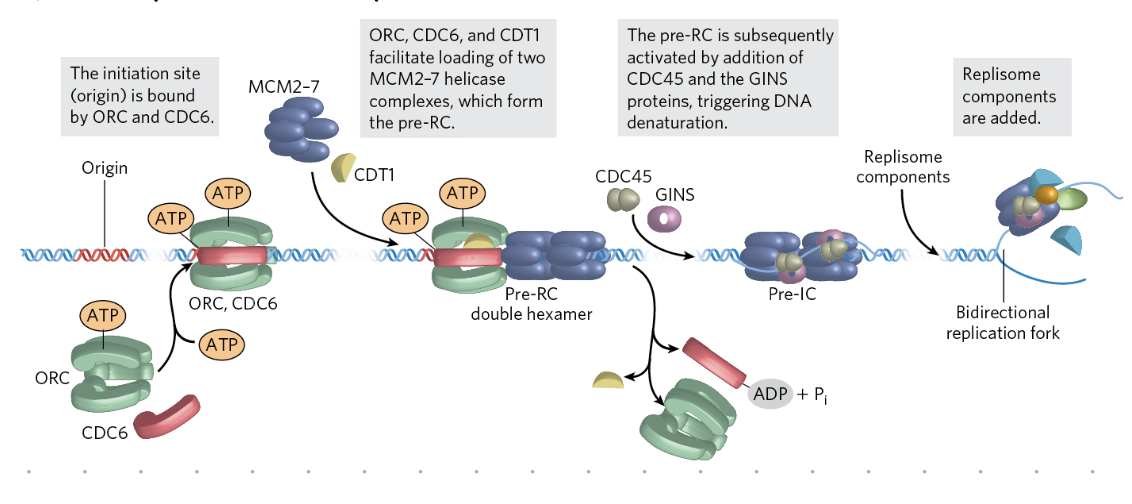
initiation of replication (eukaryotes)
G1 phase
origin sites selected
pre-replicative complexes (pre-RCs) assemble
S phase
active CDKs phosphorylate and activate pre-RCs → recruit DNA polymerases
clusters of 2 to 80 sites initiated at a time
CDKs also inhibit pre-RC formatio if replication already occured
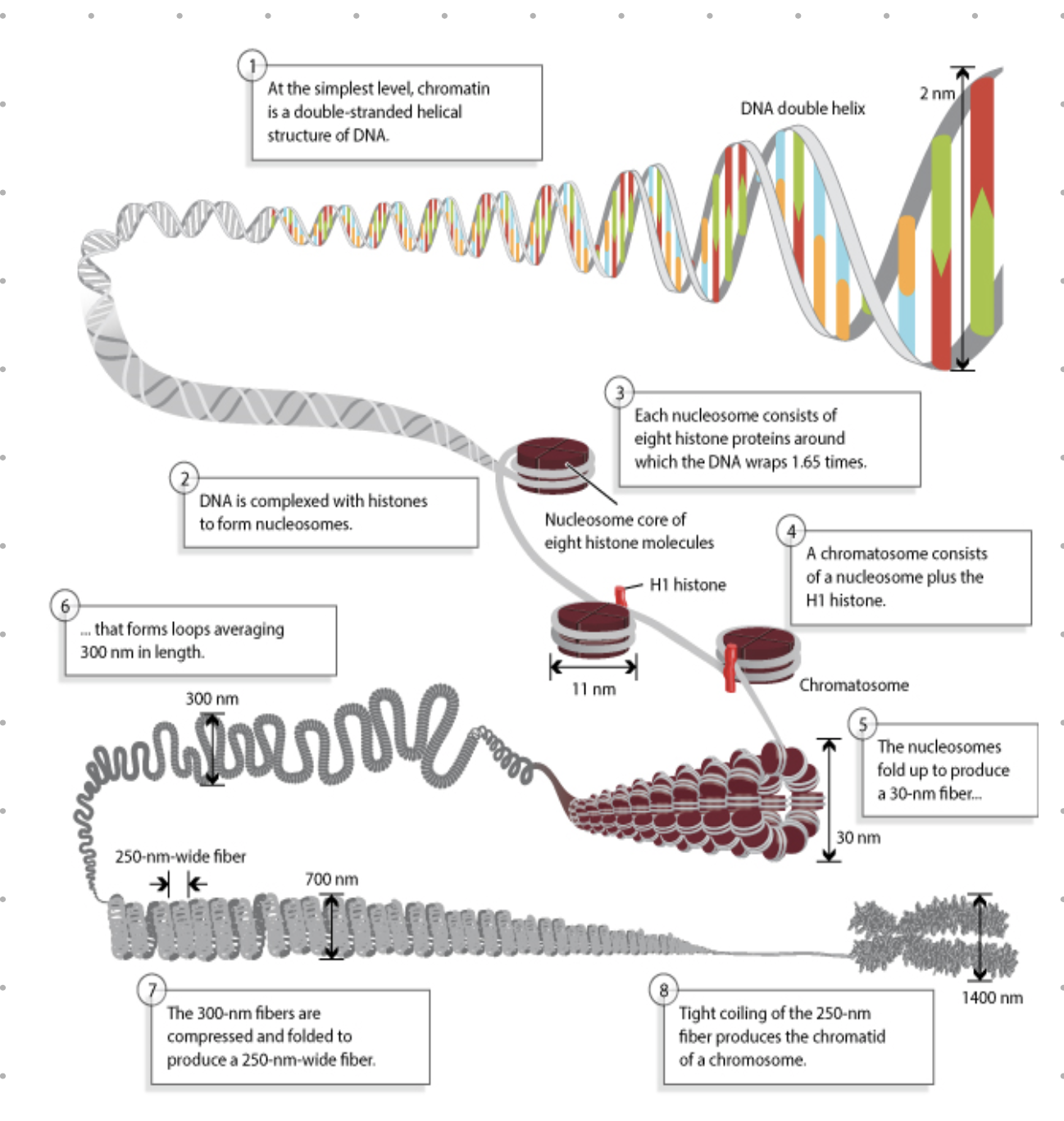
repacking chromosomes
DNA is packaged in cell - wound around histones
existing histones can be reused
replication doubles DNA → more histones needed
synthesis controlled at transcitional and post-transcriptional levels
coupled to the cell cycle (mostly during S phase)
histone synthesis
many copies of histone genes in the genome. can be quickly transcribed
hostone genes = no introns - no need to spice
hostone mRNA are not polyadenylated - mRNA does not last long
telomeres
ends of chromosomes made up of repeating sequences known as telomeres
protects genetic infor
in humans
repeat is 5’ - TTA GGG -3’
can extend as much as 10kb
double stranded except end where 3’ extends beyond 5’end
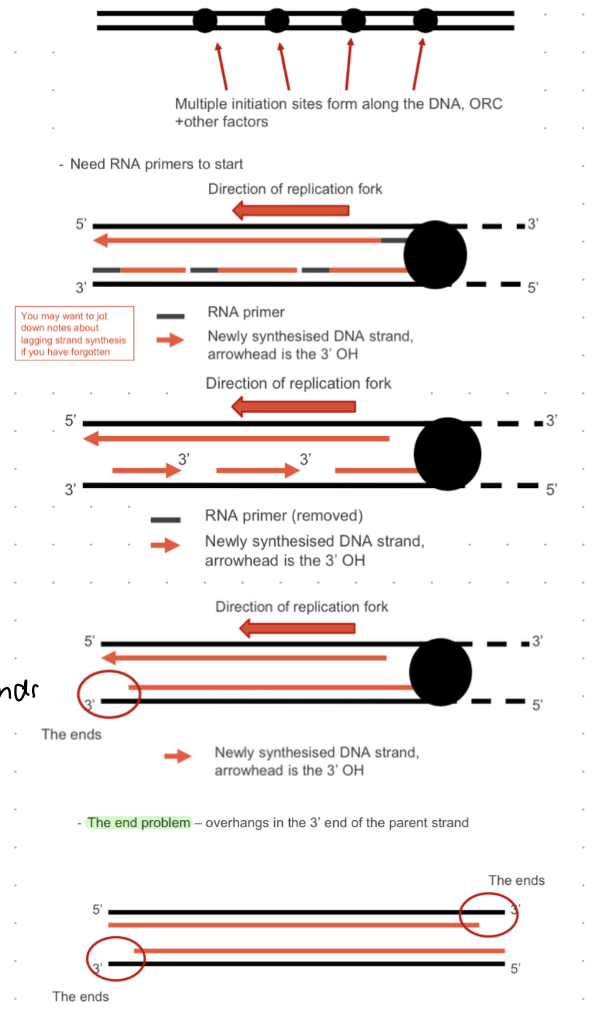
eukaryotic DNA replication - step by step
start at ORI (many sites)
RNA primers bind at ORI
DNA synthesized in 5’ direction
etc etc
end problem = overhangs in the 3’ end of the parent strand
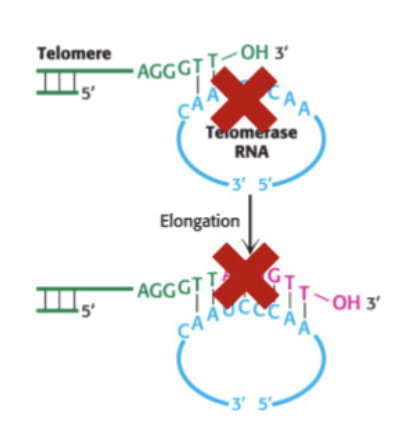
telomerase
recognises overhand and synthesizes complementary strand
low activity in somatic cells
high activity in germ line cells zygote starts with full length telomeres
high activity in highly proliferative stem cells
Telomerase, on the other hand, is the enzyme responsible for adding telomeres to the ends of the chromosome. Telomerase has a single-stranded RNA segment, which serves as a template for a single-stranded DNA. These single-stranded DNA fragments are repeated and added to the 3-prime end of the chromosome.
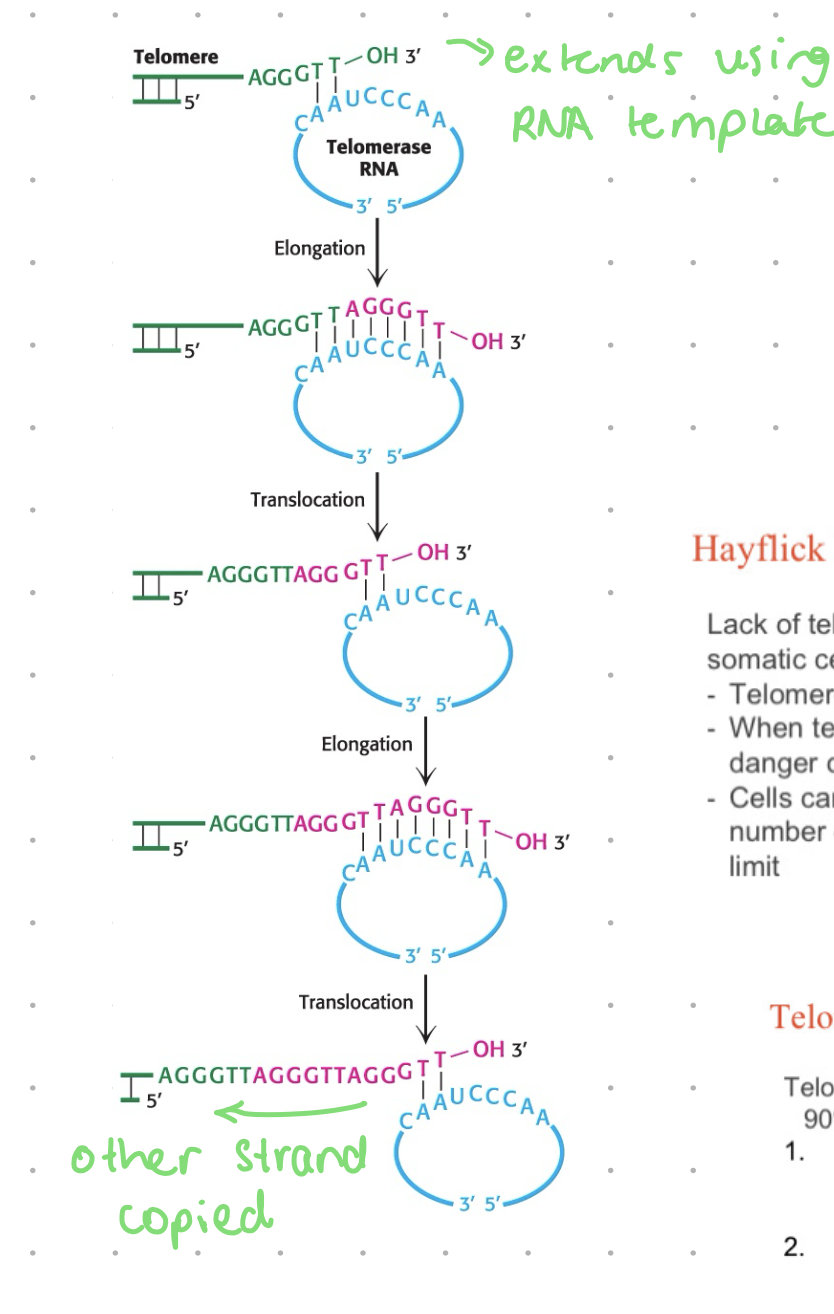
ribonucleoprotein
RNA + protein
RNA: ~1.5 copies of the complement telomere sequence - the template
protein: reverse transcriptase - DNA polymerase that uses RNA template to make a copy of DNA
5’ end extends by lagging strand mechanisms
overrhang on the 3’ end tucks in and caps the end
capping proteins protect the ends from nucleases
immortal cells
eg, cancer cells, cells continue to divide due to telomerase activity
HeLa cells were the first immortal cell line, derived from cervical cancer patient - still commonly used as a model cell line
cells which can divide indefinitely in culture
cells/genome across lifespan
human cells start w/ ~10k bp of telomeres are birth
some cells can survive an entire lifespan of replication erosion without suffering cell death (eg, those that do not divide much)
shortening telomeres could be a limiting factor in determining organism’s life span
hayflick limit
lack of telomerase activity - as seen in most somatic cells
telomeres shorten after each round of mitosis
when too short, cell is in danger of losing coding genes
cells can no longer divide (senescence) - number of times it can divide = hayflick limit
telomerases - a drug target
telomerase is active in between 80-90% of all cancers
targeting the RNA component with antisense oligodeoxynucleotides and RNaseH
reverse-transcriptase inhibitors (eg, AZT) or inhibitors of the catalytic protein subunit
dolly the sheep
cloned by taking nucleus from adult cell from mammary gland
died 203 - age 6 (LE 10-20 yrs)
shorter telomeres - suspected to be use of adult cells in cloning process
NOT CASE
no evidence of health usses related to accelerated aging
other clones had different telomere lengths + aged normally
examples of syndromes related to telomeres - so telomere length is important
copying DNA in a tube - ingrediants
template
DNA polymerase
Primers - forward + reverse
dNTPs
buffer (optimal Mg2+, pH and ionic strength)
DNA synthesis: tube vs. cell
much simpler than in cell, we can add all relevant enzymes, synthesis RNA primers that need to be removed later, make a substrate, maintain pH and ionic strength
PCR
polymerase chain reaction
PCR steps
mixutre heated to separate DNA strands - contrast to cell where helicase is responsible - heating separates weak (H bonds) forces between strands
annealing of primers - cool and force primers to anneal to sequence wanting to be amplified
extension - complementary bases hydrogen bond, polymerase catalyses formation of phosphodiester bond, adds nucleotide to growing chain (3’OH to 5’P direction)
cycling - repeat, amplicon (newly synthesized strand) becomes template for next cycle, doubling in each cycle. note in the cell, synthesis of entire genome only occurs once at a time, with PCR we can keep cycling as many times as desired