6 Enzymes
1/100
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
101 Terms
Enzymes:
A. are proteins (with few exceptions).
B. can be denatured and still retain full activity.
C. have names always ending in “-ase.”
D. are also referred to as “coenzymes.”
A. are proteins (with few exceptions).
With the exception of a few classes of catalytic RNA molecules, enzymes are proteins.
Coenzyme:
organic molecule that binds to the active sites of certain enzymes to assist in the catalysis of a reaction.
Cofactors:
is the larger group that includes ions and coenzymes.
a non-organic molecule that loosely bind to an enzyme’s active site
An apoenzyme:
A. is the nonprotein component of a holoenzyme.
B. always requires an inorganic ion for its activity.
C. requires a cofactor for its activity.
D. is an enzyme consisting of RNA rather than protein.
C. requires a cofactor for its activity.
A coenzyme or metal ion that is very tightly or even covalently bound to the enzyme protein is called a prosthetic group. A complete, catalytically active enzyme together with its bound coenzyme and/or metal ions is called a holoenzyme. The protein part of such an enzyme is called the apoenzyme or apoprotein. An apoenzyme requires a cofactor for its activity
Some enzymes require an inorganic ion for catalytic function. When this inorganic ion is very tightly or covalently bound by the enzyme it is called a(n):
A. apoenzyme.
B. prosthetic group. C. holoenzyme.
D. catalyst.
B. prosthetic group.
A coenzyme or metal ion that is very tightly or even covalently bound to the enzyme protein is called a prosthetic group.
________ is NOT an E.C. class name for enzymes.
A. Transferases
B. Polymerases
C. Lyases
D. Isomerases
B. Polymerases
Polymerases is not one of the 7 E.C. classes of enzymes.
ground state
starting point for either the forward or reverse reaction
transition state (‡)
the point at which decay to substrate or product are equally likely
biochemical standard free- energy change ∆G′°
the standard free-energy change at pH 7.0
activation energy (∆G‡)
difference between the ground state energy level and the transition state energy level
active site
providesa specific environment in which a given reaction can occur more rapidly
substrate
the molecule that is bound to the active site and acted upon by the enzyme
simple enzymatic reactions can be written as
E + S ⇌ ES ⇌ EP ⇌ E + P
What are E, S, and P in the simple enzymatic reaction?
the enzyme, substrate, and product
What are ES and EP in the simple enzymatic reaction?
transient complexes of the enzyme
What is the free-energy starting point for a reverse reaction designated as?
A. transition state (‡)
B. ground state
C. biochemical standard free energy
D. activation energy (∆G‡)
B. ground state
The starting point for either the forward reaction or the reverse reaction is called the ground state, the contribution to the free energy of the system by an average molecule (S or P) under a given set of conditions.
Caatlysts do what to the Activation Energy and what to the reaction reate?
lower and Increase
reaction intermediate
any species on the reaction pathway that has a finite chemical lifetime
– example: ES and EP complexes
rate-limiting step
the step in a reaction with the highest activation energy that determines the overall rate of the reaction
activation energies are what to chemical reactions?
barriers
true or false. Catalysts Do Not Affect Reaction Equilibria
true
true or false. enzymes are used up in the process
false
true or false. any enzyme that catalyzes the reaction S → P also catalyzes the reaction P → S
True
The conversion of sucrose to CO2 and water has a very large and negative ∆G′°. Why is the conversion NOT spontaneous?
A. Only some reactions have a negative ∆G′°.
B. Conversion requires heat and O2.
C. Conversion requires the presence of cofactors.
D. There is a very large activation energy barrier.
D. There is a very large activation energy barrier.
The difference between the ground state energy level and the transition state energy level is the activation energy (∆G‡). The rate of a reaction reflects this activation energy: a higher activation energy corresponds to a slower reaction. Activation energies are energy barriers to chemical reactions. Without such energy barriers, sucrose would spontaneously form CO2 and water.
What does an enzyme change relative to an uncatalyzed reaction?
A. the equilibrium constant
B. the rate of the reaction
C. the pH
D. the free energy change of the reaction
B. the rate of the reaction
The role of enzymes is to accelerate the interconversion of S and P. The enzyme is not used up in the process, and the equilibrium point is unaffected. However, the reaction reaches equilibrium much faster when the appropriate enzyme is present because the rate of the reaction is increased.
reaction equilibria are linked to the…
standard free-energy change for the reaction, ∆G′°
reaction rates are linked to the…
activation energy, ∆G‡
binding energy, ∆GB
energy derived from noncovalent enzyme-substrate interaction
mediated by hydrogen bonds, ionic interactions, and the hydrophobic effect
major source of free energy used by enzymes to lower the activation energy
covalent interactions between enzyme and substrate do what to the activation energy?
lower
“lock and key” hypothesis
enzymes are structurally complementary to their substrates
– would make for a poor enzyme!
the full complement of interactions between substrate and enzyme is formed only when…
the substrate reaches the transition state
How does the concept of “induced fit” support the current theory of substrate-enzyme interaction?
A. Enzyme structure is rigid, allowing for competition for the active site and easy ejection of the final product.
B. Enzyme conformational changes demonstrate feedback regulation of the enzyme and the ability to “turn off” activity.
C. Having a rigid structure allows the enzyme to bind the substrate and stabilize the ES complex.
D. Enzyme conformation changes allow additional stabilizing interactions with the transition state and enhance the activity.
D. Enzyme conformation changes allow additional stabilizing interactions with the transition state and enhance the activity.
Some weak interactions are formed in the ES complex, but the full complement of such interactions between substrate and enzyme is formed only when the substrate reaches the transition state. Enzyme conformational changes allow for additional stabilizing interactions between the enzyme and the transition state.
the sum of the unfavorable activation energy ∆G‡ and the favorable binding energy ∆GB results in…
a lower net activation energy
weak binding interactions between the enzyme and the substrate
drive enzymatic catalysis
Binding energy is defined as the:
A. energy derived from noncovalent enzyme-substrate interaction.
B. energy derived from release of the product and release from the EP interaction.
C. energy released from the conversion of ES to EP.
D. difference in the uncatalyzed activation energy and
catalyzed activation energy.
A. the energy derived from noncovalent enzyme-substrate interaction.
The energy derived from noncovalent enzyme-substrate interaction is called binding energy, ∆GB. Binding energy is a major source of free energy used by enzymes to lower the activation energies of reactions.
optimized binding energy in the transition state is accomplished by positioning a substrate in a
cavity (the active site), removed from H2O
specificity
ability to discriminate between a substrate and a competing molecule – given by binding energy
entropy reduction
large restriction in the relative motions of two substrates that are to react
Activation energy ∆G‡ includes:
-the entropy of molecules in solution
– the solvation shell of hydrogen-bonded water that surrounds and stabilizes most biomolecules in aqueous solution
– the distortion of substrates that must occur in many reactions
– the need for proper alignment of catalytic functional groups on the enzyme
desolvation
replacement of the solvation shell of structured water around the substrate with weak bonds between substrate and enzyme
– replaces most or all hydrogen bonds between the substrate and H2O
substrate must undergo… to cause an unfavorable free-energy change
distortion. binding energy compensates thermodynamically for this
induced fit
mechanism by which the enzyme itself undergoes a conformational change when the substrate binds, induced by multiple weak interactions with the substrate
– enhances catalytic properties
What is the primary source of the energy enzymes used to reduce activation energies?
A. desolvation of the substrate
B. kinetic energy from the transition state
C. noncovalent enzyme-substrate interaction
D. covalent enzyme-product interaction
C. noncovalent enzyme-substrate interaction
Weak binding interactions between the enzyme and the substrate provide a substantial driving force for enzymatic catalysis.
How does the induced fit mechanism of enzyme catalysis work?
A. The enzyme assumes a conformation identical to the substrate.
B. The enzyme undergoes a conformational change to maximize weak interactions to the substrate.
C. The substrate binds the active site of the enzyme.
D. The enzyme undergoes entropy reduction to accommodate
substrate.
B. The enzyme undergoes a conformational change to maximize weak interactions to the substrate.
The enzyme usually undergoes a change in conformation when the substrate binds, induced by multiple weak interactions with the substrate, a mechanism referred to as induced fit. Induced fit brings specific functional groups on the enzyme into the proper position to catalyze the reaction and permits formation of additional weak bonding interactions in the transition state.
Which statement about enzymes is false?
A. They lower the activation energy of a reaction.
B. They alter the overall thermodynamics of a reaction.
C. A substantial amount of their catalytic power results from
binding of the substrate(s) through weak interactions.
D. They increase the rates of reactions by 105-fold to 1017- fold.
B. They alter the overall thermodynamics of a reaction.
Catalysts do not affect the overall thermodynamics of a reaction. Instead, catalysts increase the rate of the reaction.
catalytic functional groups aid in the cleavage and formation of bonds by a variety of mechanisms (which are):
general acid-base catalysis
covalent catalysis
metal ion catalysis
…are transferred between an enzyme and a substrate or intermediate
protons
specific acid-base catalysis
uses only the H+ (H3O+) or OH– ions present in water
general acid-base catalysis
mediated by weak acids or bases other than water
Which amino acid might be expected to have the LEAST effect on the function of an enzyme if it replaces a Glu residue in the enzyme?
A. Gly
B. Lys
C. His
D. Asp
D. Asp
Asp and Glu have similar general acid forms (R—COOH) and general base forms (R—COO–). Thus, replacing a Glu residue with an Asp residue would have a lesser effect on the function of the enzyme than a Gly, Lys, or His residue.
covalent catalysis
transient covalent bond forms between the enzyme and the substrate
– catalysis only results when the new pathway has a lower activation energy than the uncatalyzed pathway
– all new steps must be faster than the uncatalyzed reaction
Metal Ion Catalysis
help orient the substrate for reaction
stabilize charged reaction transition states
mediate oxidation-reduction reactions by reversible changes in the metal ion’s oxidation state
An enzyme accepts H+ from hydronium and transfers it to an amine group of the substrate. The result is an increase in the rate of release of the product. This is an example of:
A. metal ion catalysis.
B. general acid catalysis.
C. covalent catalysis.
D. specific acid catalysis.
B. general acid catalysis.
General acid-base catalysis refers to proton transfers mediated by weak acids and bases other than water. Thus, an enzyme-mediated transfer of a H+ from hydronium to an amine group of a substrate is an example of general acid catalysis.
How to Assess Enzymatic Activity
Disappearance of substrate • Appearance of product
Ex. of enzymatic Activity
Appearance of a colored product
Appearance of a UV absorbent product
Appearance of a radioactive product
S+E↔P+ E
When using the above reaction, we can express the rate based on:
A. Substrate concentration
B.Product concentration
C.Enzyme concentration
D.Using either the substrate or the product concentration
E.Using either the substrate, product, or enzyme concentration
D.Using either the substrate or the product concentration
Michaelis-Menten Plot (saturation plot)
shape is hyperbolic – a characteristic of many enzymes
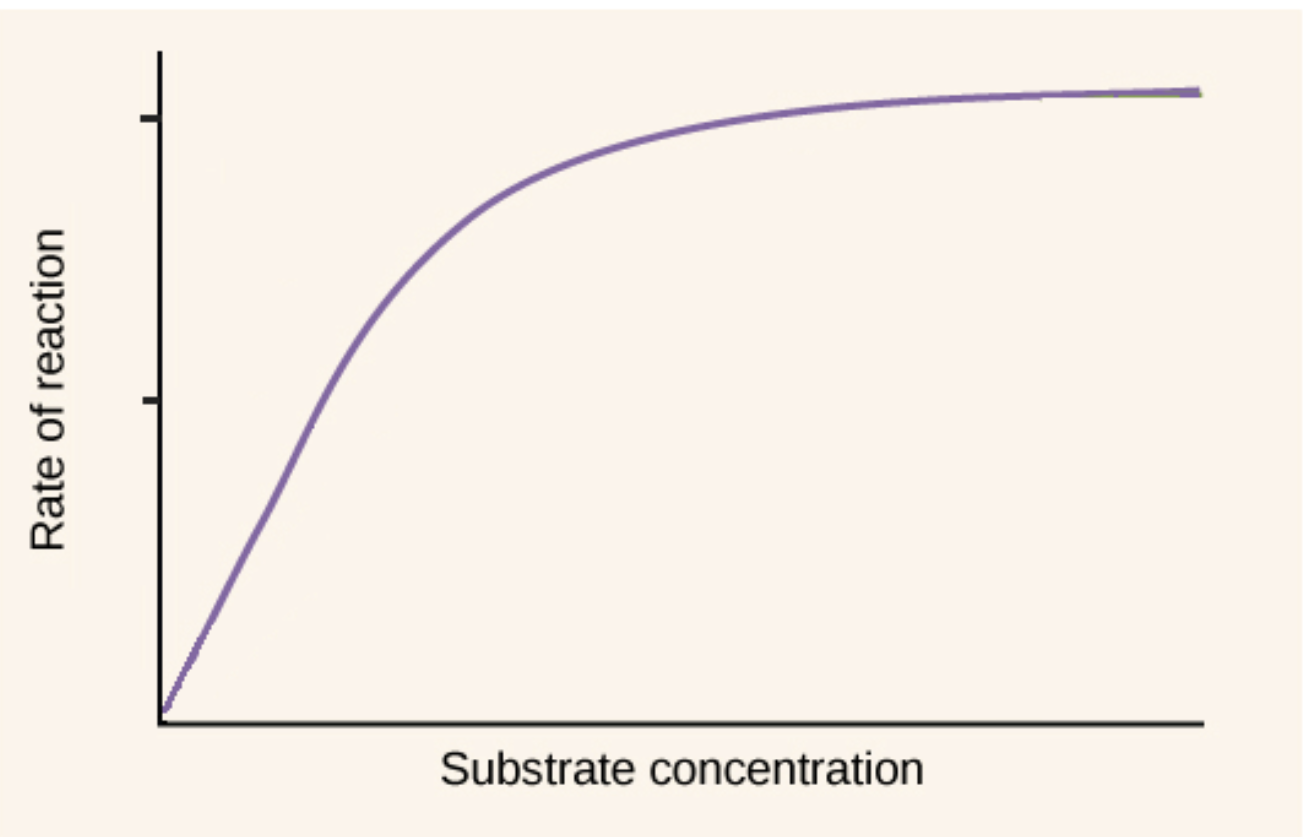
Vmax is observed when…
virtually all the enzyme is present as the ES complex
– further increases in [S] have no effect on rate
– responsible for the plateau observed
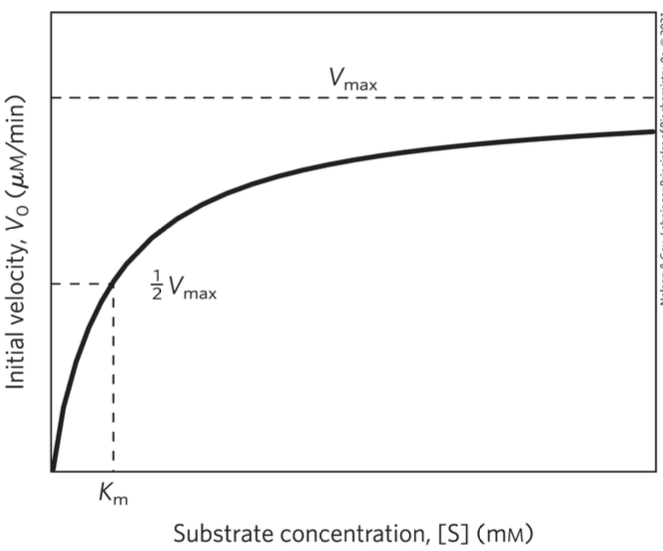
Km
this is the concentration of substrate needed so the reaction goes at half the maximum speed (Vmax)
Quantitative Expression of Enzyme Behavior. Rate constant:
S + E (top K1, bottom K-1)↔ ES (top K2) → P +E
True or false. S + E (top K1, bottom K-1)↔ ES (top K2) → P +E. The reverse reaction (P → S) is not considered because the equation describes the initial rates when [P] is near zero
true
True of False. S + E (top K1, bottom K-1)↔ ES (top K2) → P +E. The ES complex is not a STEADY STATE INTERMEDIATE
false
true or false. S + E (top K1, bottom K-1)↔ ES (top K2) → P +E. The concentration of ES remains relatively constant because it is produced and broken down at the same rate.
true
Rate of ES production =
Rate of ES breaking down. k1[S][E] = k-1[ES] + k2[ES]
Vmax
the maximum rate that can be observed in the reaction
– Substrate is present in excess
– Enzyme can be saturated (zero order reaction)
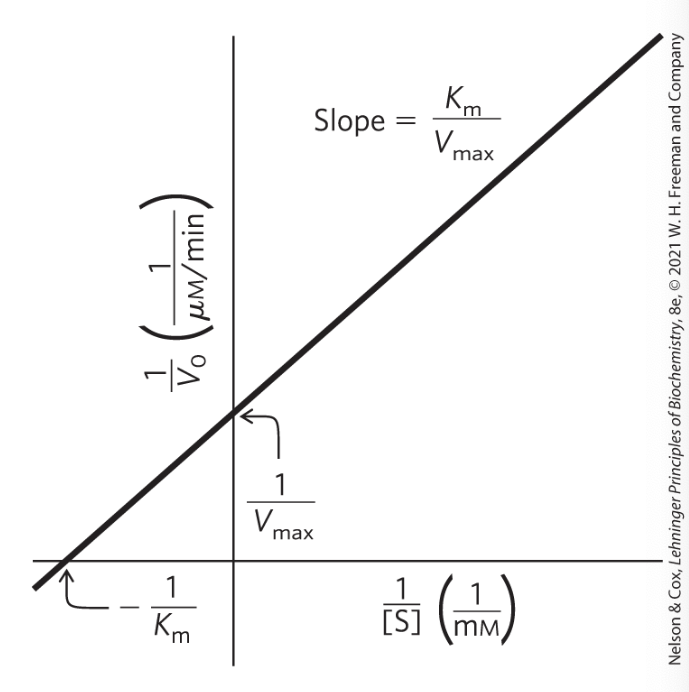
A Double-Reciprocal, or Lineweaver- Burk, Plot. used for?
for enzymes obeying the Michaelis- Menten relationship, a plot of 1/V0 versus 1/[S] yields a straight line
enzyme inhibitors
molecules that interfere with catalysis, slowing or halting enzymatic reactions
2 classes of enzyme inhibitors:
reversible and irreversible
types of reversible inhibition:
competitive inhibition
uncompetitive inhibition
mixed inhibition
noncompetitive inhibition
competitive inhibitor:
competes with the substrate for the active site of an enzyme
type of reversible inhibition
binds only to free enzyme. Often this binding event occurs on the active site of the target, precisely where substrate also binds. Although this is the case for a majority of competitive inhibitors, it is a misleading oversimplification. It is more appropriate to state that the binding of a competitive inhibitor and the binding of substrate are mutually exclusive events.
competitive inhibition on Km and Vmax
Raise apparent Km, no change to Vmax
Noncompetitive Inhibitor
Binds away from active site
• Causes an allosteric change—a shape change
• Has equal affinity for empty enzyme or enzyme bound to substrate
The reaction can never reach its normal Vmax regardless of how much substrate we add. A subset of the enzyme molecules will always be “poisoned” by the inhibitor
noncompetitive inhibition on Km and Vmax
No change to Km, lowers Vmax
uncompetitive inhibitor:
binds at a site distinct from the substrate active site
unlike a competitive inhibitor, binds only to the ES complex
type of reversible inhibition
uncompetitive inhibition on Km and Vmax
Lowers apparent Km, lowers Vmax
mixed inhibitor:
binds at a site distinct from the substrate active site
binds to either E or the ES complex
type of reversible inhibition
mixed inhibition on Km and Vmax
Affects apparent Km, lowers Vmax
suicide inactivator = mechanism-based inactivators =
undergo the first few steps until it is converted into a compound that combines irreversibly with the enzyme class of irreversible inhibitors
transition-state analogs
stable molecules designed to resemble transition states
type of irreversible inhibitors
bind to an enzyme more tightly than does the substrate in the ES complex
enzyme kinetics
the discipline focused on determining the rate of a reaction and how it changes in response to changes in experimental parameters
the curve expressing the relationship between [S] and V0 can be expressed by the Michaelis-Menten equation:
V0 = "Vmax [S]" /"Km + [S] "
![<p><span><em><span>V</span></em><sub><span>0 </span></sub><span>= "V</span><sub><span>max </span></sub><span>[S]</span></span><span style="font-family: "Cambria Math";"><span>" /"</span></span><span><span>K</span><sub><span>m </span></sub><span>+</span><sub><span> </span></sub><span>[S] </span></span><span style="font-family: "Cambria Math";"><span>"</span></span></p>](https://knowt-user-attachments.s3.amazonaws.com/e4f5c881-f683-4135-8257-29ae9ae5b2ec.png)
for a two-step Michaelis-Menten mechanism,
Vmax = k2[Et]
general rate constant, kcat turnover number
describes the limiting rate of any enzyme-catalyzed reaction at saturation
if one step in a multistep reaction is clearly limiting, kcat equals the rate constant for the step
more complex when several steps are rate-limiting
, kcat = Vmax/[Et]:
The rate-limiting step of an enzyme-catalyzed reaction:
A. is always the last step.
B. has a rate constant equal to kcat.
C. is always the fastest step.
D. is always slower than the rate of diffusion of the substrate to
the active site.
B. has a rate constant equal to kcat.
The general rate constant, kcat, describes the limiting rate of any enzyme-catalyzed reaction at saturation.
specificity constant
the rate constant for the conversion of E + S to E + P
true or false. Michaelis-Menten steady-state kinetics can distinguish between pathways that have a ternary intermediate and pathways that do not
true
Parallel Lines Indicate a what Pathway?
a Ping-Pong (Double-Displacement)
Enzyme Activity Depends on
the pH range over which an enzyme undergoes changes in activity can provide a clue to the type of amino acid residue involved
protease
an enzyme that catalyzes the hydrolytic cleavage of peptide bonds
acylation phase
the peptide bond is cleaved and an ester linkage is formed between the peptide carbonyl carbon and the enzyme
deacylation phase
the ester linkage is hydrolyzed and the nonacylated enzyme is regenerated
retrovirus
possess an RNA genome and an enzyme, reverse transcriptase, that uses RNA to direct the synthesis of a complementary DNA
example = HIV
peptidoglycan
major component of the bacterial cell wall
consists of polysaccharides and peptides cross-linked in several steps that include a transpeptidase reaction
β-lactamases =
enzymes that cleave β-lactam antibiotics
regulatory enzymes
catalytic activity increases or decreases in response to certain signals
allows the cell to meet changing needs for energy and biomolecules
activities of regulatory enzymes are modulated in a variety of ways:
allosteric enzymes = function through reversible, noncovalent binding of regulatory compounds called allosteric modulators or allosteric effectors (small metabolites or cofactors)
reversible covalent modification
binding of separate regulatory proteins
removal of peptide segments by proteolytic cleavage
homotrophic =
regulation in which the substrate and modulator are identical
heterotropic =
regulation in which the modulator is a molecule other than the substrate
Allosteric enzymes:
A.are never regulated by substrate binding.
B.have their activity changed by changes in intersubunit interactions.
C.exhibit Michaelis-Menten kinetics.
D.always have both inhibitory and activating modulators.
B. have their activity changed by changes in intersubunit
interactions.
Allosteric proteins are those having “other shapes” or conformations induced by the binding of modulators. Conformational changes induced by one or more modulators interconvert more-active and less-active forms of the enzyme.
zymogen
inactive precursor that is cleaved to form an active protease enzyme