Fragment Analysis
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
14 Terms
What is fragment analysis
Fragment analysis = the study of different sized pieces of DNA, usually to identify a region of the genome or diagnose disease
- In the clinical world, it usually means that the fragment sizes are analyzed using capillary electrophoresis
o Can be done using the type of instrument that can do sanger sequencing
What is capillary electrophoresis (CE)
Capillary electrophoresis (CE)
- Separates DNA by size (like other types of nucleic acid electrophoresis)
- The “gel” is called a polymer and is often polyacrylamide based which has a high resolution
- The polymer is inside a thin tube called a capillary
o Most instruments have a least 4 capillaries and 8 capillaries is very common, some can have up to 96
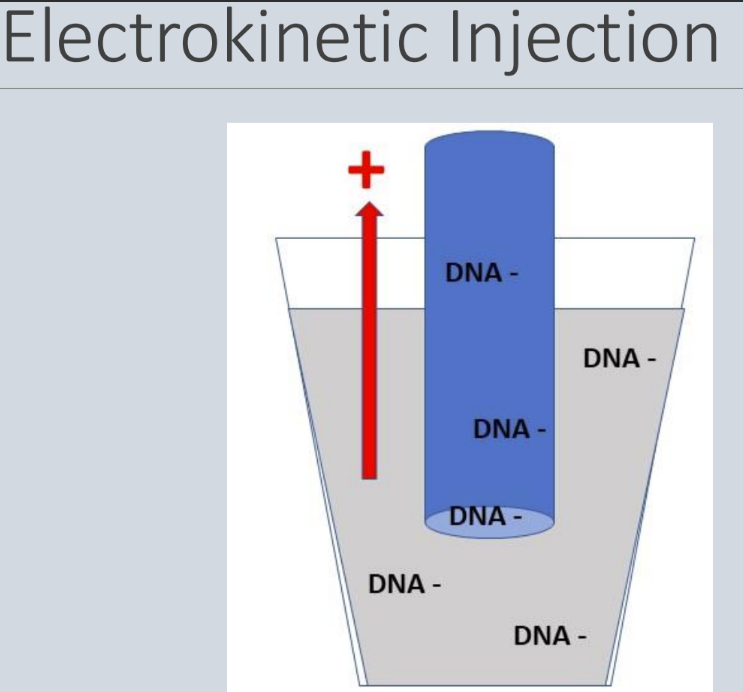
- Each sample enters a capillary by electrokinetic injection and a high voltage is applied, forcing the negatively charged DNA through the capillary (small fragments go first, followed by the slow big fragments)
o Size relates to speed
- The DNA (which must be fluorescently labeled) passes the path of a laser beam. The dye signals are separated by a diffraction system and the CCD camera detects the different fluorescence
List the applications of fragment analysis and provide examples of molecular characteristics that can be detected by fragment analysis
Microsatellite (STR) analysis (most common)
- Human identity testing (forensics)
- Pathogen typing
- Microsatellite instability (like colon cancer)
- Loss of heterozygosity
- Genetic diagnosis in repeat disorders (like FXS)
Other applications
- SNP genotype using specialized PCR kits
- Amplified fragment length polymorphism (AFLP)
- Quantification using relative fluorescence
o Multiplex ligation-dependent probe amplification (MLPA)
o Copy number variation
What is multiplexing?
Multiplexing = allows to see many different products at the same time
- Each dye in a reaction emits light at a different wavelength when excited by the laser
- An advantage of capillary electrophoresis is that the DNA can be labeled with different fluorescent tags and thus even fragments of the same size can be distinguished apart within the same capillary (multiplexing)
- Multiplexing can ...
o Occur during PCR, followed by CE
o Or PCR reaction can be performed separately and then “pooled” together prior to CE
§ Simpler approach because then PCR does not have to be optimized for multiple primers in the same reaction
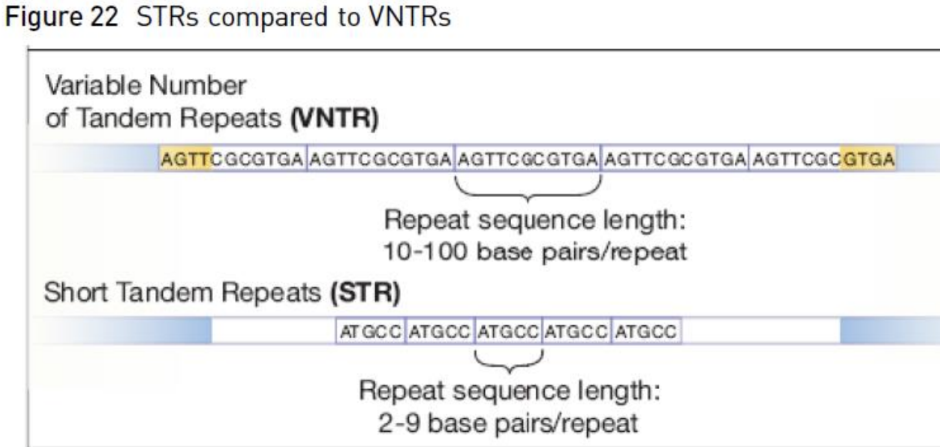
What is a microsatellite
Microsatellite = Short Tandem Repeats (STRs)
- Are polymorphic regions of DNA with repeated nucleotide sequences
- Most STRs have repeat units of 2-7 nucleotides in length
o SMALLER REPEAT UNITS
- Ex = CTA CTA CTA CTA
o This DNA has 4 repeats of nucleotides in length (trinucleotide repeat)
What is a minisatellite
Minisatellites = Variable Number Tandem Repeats (VNTRs)
- Repeat regions that have larger units of nucleotides that are repeated
- Usually about 10-100 nucleotide repeating units (motifs)
o BIGGER REPEAT UNIT
What is the difference between a micro and minisatellite
Difference = the number of nucleotides per unit (how big is the unit that gets repeated)
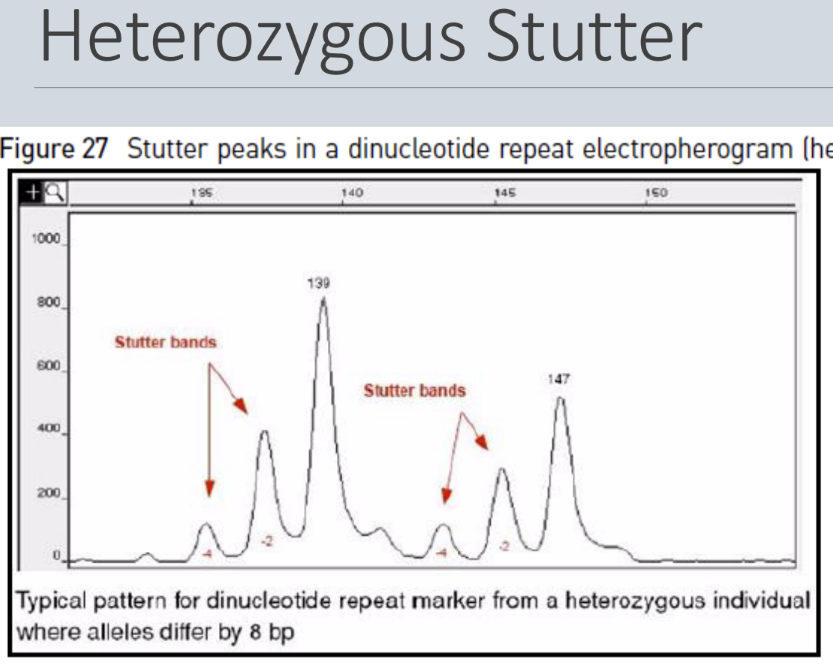
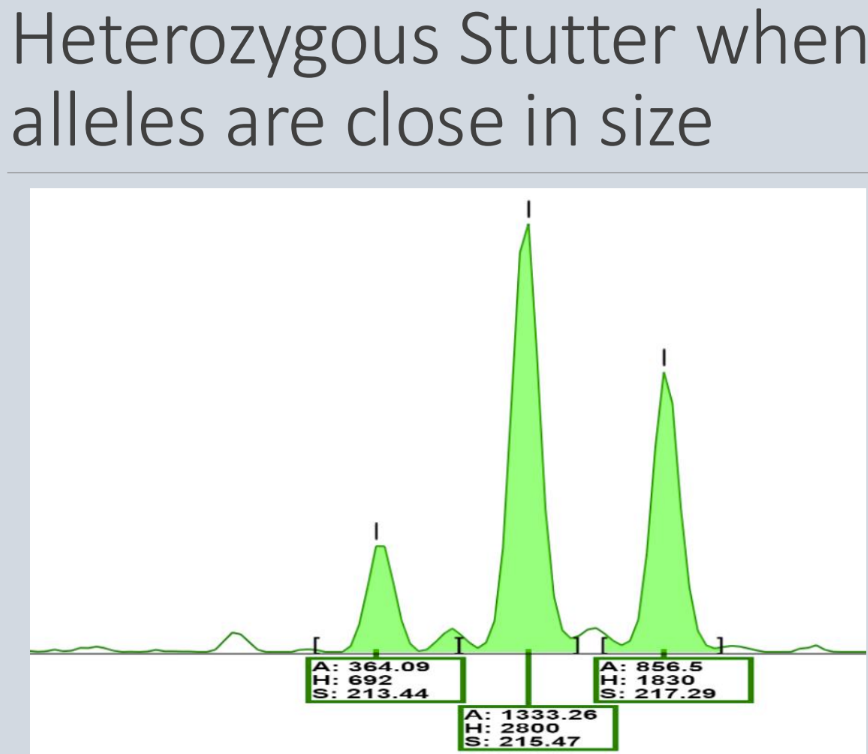
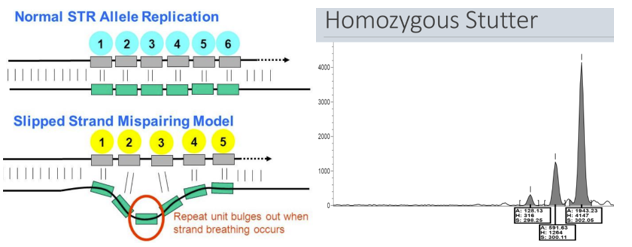
What is a stutter
Stutter = minor products that are 1-4 repeat units shorter are often produced during PCR amplification of short STRs
- It tends to happen most in the short STRs, such as dinucleotide, trinucleotide, and tetranucleotide repeats
- It's thought that stutter is caused by slippage of polymerase during extension
What is the workflow of STR genotyping by fragment analysis
Workflow of STR genotyping by fragment analysis
1. Isolate DNA
2. Purify DNA if necessary
3. Quantify DNA
4. PCR amplification using dye-labeled primers
5. Capillary electrophoresis
6. Data analysis
How does STR genotyping work with fragment analysis
STR genotyping
- Different STR alleles can be distinguished by the size of the PCR product
- The high resolution of capillary electrophoresis allows for different numbers of repeats of only a few nucleotides in length to be distinguished
- There may be many different STR alleles at a particular locus
o Ex: an STR locus may be a trinucleotide repeat with a possibility of 12, 13, or 14 repeats
o OR there could be many more...normal individuals may range from 10-20 repeats at a particular locus while individuals affected by a genetic disorder may have 50-100+ repeats at that locus
o Each locus is different

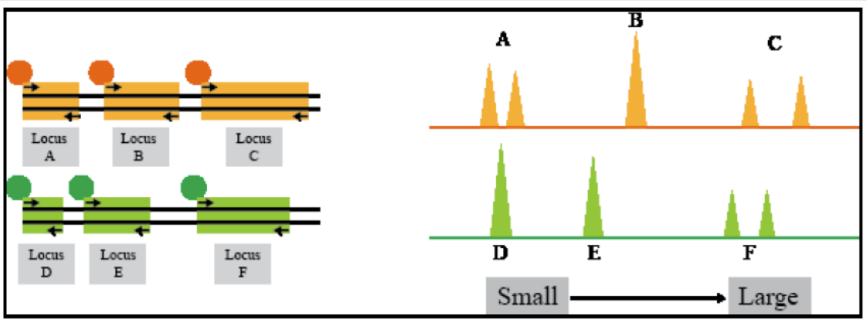
How do you do STR analysis for multiple loci
STR analysis for multiple loci
- Look at multiple loci at one time using different fluorescent tags (where fragment analysis can really shine)
- Method used in human identity testing (paternity and forensics)
- Each locus doesn’t need it’s own tag, because as long as it’s far enough apart in size (alleles don’t overlap), then several loci can share a color
- Fragments are separated by size AND fluorescent dye to identify them
What can be determined from CE fragment analysis
Fragment analysis by CE can be used to determine...
1. Size of the DNA fragment = based on time it takes to travel through the capillary
2. Quantity = based on the height of the peak (signal intensity)
3. Genotype = based on the product sizes of different alleles
The height = intensity (more fragments of that size) The biggest fragment = far right with the most bases (bigger because takes longer for it to go through the capillary)
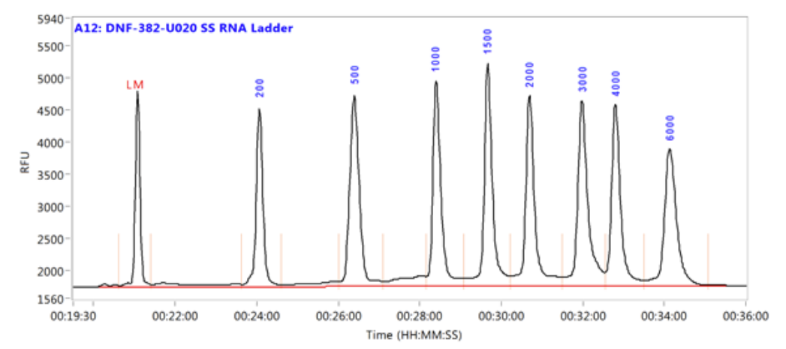
What is a size standard
Size standards = ladders
- The interpretation of fragment size is dependent on the presence of a size standard
- Is basically a molecular weight marker or ladder
- It is run in the same capillary as the sample (pooled) and must have a fluorescent marker that is different from the sample
By having a size standard in each sample, there is a correction for small variations in mobility between capillaries and samples due to salt concentration, pH, temperature, etc
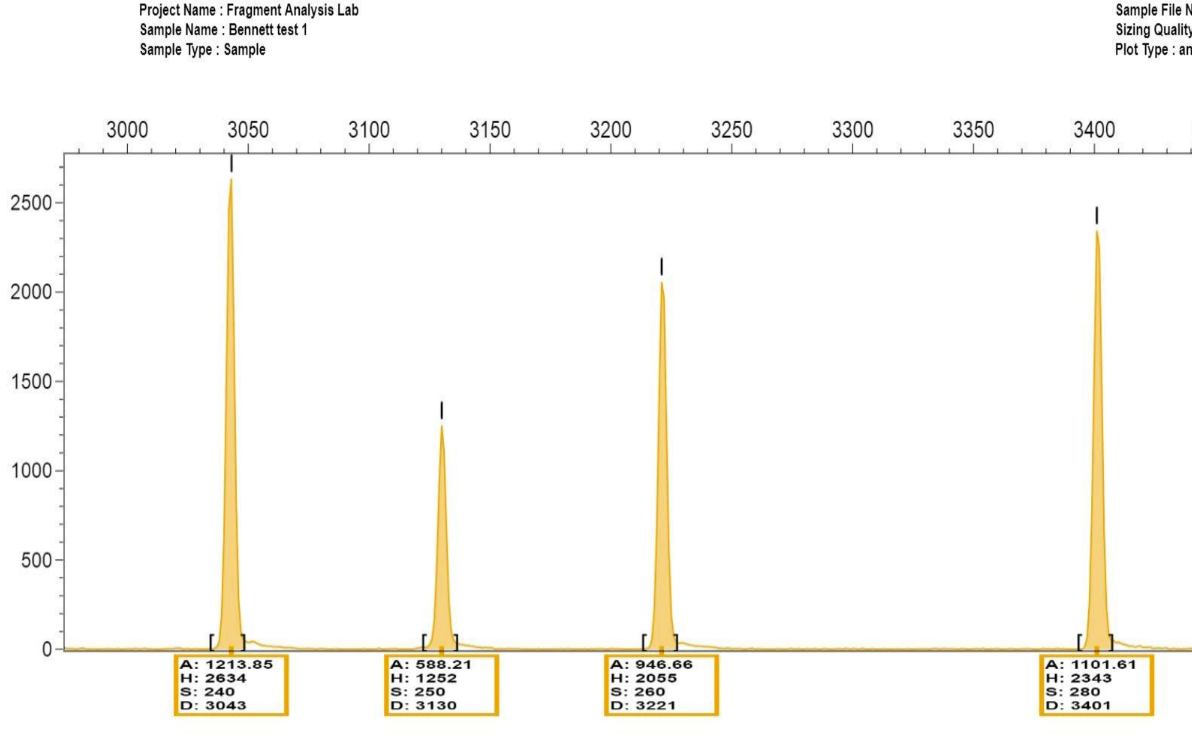
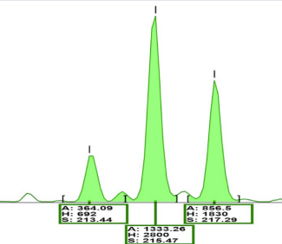
What is shown in the image? Which ones are the true alleles
Far right 2 are the true