Quest 2
1/130
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
131 Terms
acidophile
bacteria with an optimal pH below 7
alkaliphile
bacteria with optimal pH above 7
ampicillin
beta-lactam ring antibiotic that inhibits cell wall transpeptidase for both gram-positive and gram-negative bacteria
bacitracin
antibiotic that prevents the recycling of bactoprenol into the cytoplasm after it released the peptidoglycan precursor, trapping it in outside the plasma membrane
bactoprenol
lipid carrier molecule that carries peptidoglycan monomers through the plasma membrane, and must recycle back into the cell
chaperone proteins
help bind proteins together in extreme high temperatures
compatible solutes (glycerine-betaine, proline, sucrose)
useful in halophiles or drought-tolerant bacteria, held within cells to keep equilibrium with their environment
cytostatic
antibiotic that stops cells from reproducing but does not kill them
cytotoxic
antibiotic that kills cells
DNA-binding proteins
cationic proteins used to stabilize the structure of DNA to increase stability in extreme high temperatures
halophile
salt-loving bacteria, thriving in environments with high solute concentrations
hyperthermophilic
bacteria that thrive in really hot environments. rRNA often have more G-C base pairs (3 H-bonds between bases) than A-U base pairs (2 H-bonds)
lipid bilayer vs monolayer
lipid bilayers are more fluid than a biphytanyl monolayer. means that bilayers are better at low temps, and monolayers are better at high temps.
mesophilic
bacteria that grow best at moderate temperatures
protein structure
proteins made up of multiple polypeptide chains folding onto each other to form a functional unit. nonideal conditions can denature proteins by altering the folding, rendering them nonfunctional
psychrophilic
bacteria that thrive in cold conditions
rRNA structure
made up of a large 50S ribosomal subunit (made up of 5S and 23S subunits), and a small 30S subunit (16S subunit)
saturated vs cis unsaturated fatty acids
saturated fatty acids pack tightly, ideal for high temperature, remain rigid. cis unsaturated fatty acids pack loosely, ideal for colder temperature, remain fluid.
thermophilic
bacteria that thrive in relatively high temperatures.
vancomycin
“last resort” antibiotic, binds to 2 terminal amino acids to prevent transpeptidation. few bacteria have developed resistance.
16S rRNA
rRNA found within small 30S ribosomal subunit, containing an anti-Shine Dalgarno sequence which allows it to bind to the Shine Dalgarno sequence on mRNA. it also contains a translation termination sequence. this subunit is highly conserved, and found across a variety of species.
2’ vs 3’ hydroxyl (OH)
RNA has 2’ OH, DNA does not. this makes RNA unstable due to proximity to the 3’ OH/phosphate backbone, and capacity for hydrolysis/degradation. both RNA and DNA have 3’ OH, which allows monomers to form phosphate backbones.
amino-acyl tRNA synthetase
enzyme responsible for attaching the correct amino acid to the correct tRNA
anticodon
series of three nucleotides on a tRNA molecule that correspond to the three base pairs of RNA called a codon.
codon (specific codons: start and stop)
three nucleotides of RNA or DNA that encode for a specific amino acid. Start codon: AUG (N-formylmethionine) begins amino acid sequence. Stop codons (UAA, UAG, UGA) signal ribosomes to stop translation.
gene expression
DNA is transcribed into mRNA, and is then translated into proteins.
non-template strand
DNA strand that has the same nucleotide sequence as the intended RNA, thus it is not used to form a complementary RNA strand.
palindrome
DNA sequence that reads the same 3’ to 5’ on one strand, and 5’ to 3’ on the corresponding strand.
promoter
DNA sequence of nucleotides upstream of a gene where RNA polymerase binds and unwinds the DNA. important signal for transcription.
release factor
protein useful in the end of translation that enter’s the A site of the ribosome, and cleaves the final bond between the final amino acid and the tRNA holding it.
RNA polymerase
enzyme that unwinds DNA and creates a complementary strand of RNA. contains a sigma factor protein with specificity to a promoter sequence. synthesizes RNA from 3’ to 5’. prokaryotes have 1 RNA polymerase, while eukaryotes have 3.
Shine-Dalgarno sequence
sequence of ribonucleotides signaling for ribosomes to bind, upstream of start codon. (GGAGG). complementary to 16S rRNA anti-Shine-Dalgarno sequence.
sigma factor
protein within RNA polymerase that helps it bind to a specific promoter sequence on the DNA so it can latch on upstream of the start codon and begin transcription.
streptomycin
antibiotic that binds to the 30S subunit of ribosomes, preventing protein synthesis. leads to cell death.
template strand
transcription initiation site
DNA strand used to create a complementary RNA sequence in the process of transcription. This RNA sequence would contain codons likely encoding a sequence of amino acids.
transcriptional termination site
site on DNA where RNA polymerase detaches, and becomes free to transcribe another gene.
untranslated regions (UTR)
sections of mRNA existing before the start codon or after the stop codon, such that they are transcribed but are not translated to form proteins.
ribosome structure
made up of two subunits. large (50S), and small (30S) subunits. the 30S attaches first according to the 16S subunit’s anti-shine-dalgarno sequence
use of Ecoli lysate to understand genetic code
E coli lysate (cytoplasm contents) plus DNase were used to experimentally determine the genetic code by inserting radiolabeled amino acids, and synthesized RNA sequences, scientists were able to determine which RNA sequences encoded which amino acids.
signal recognition particle
RNA-protein complex that recognizes signals in proteins and inserts the protein into the membrane
SecA
carries proteins to be secreted from the plasma membrane.
Carboxyl ( C ) vs Amino ( N ) terminus
the N terminus contains amine group, and is the seen at the starting end of a polypeptide chain. the C terminus contains a carboxyl group, is present at the end of a polypeptide chain, and is where new amino acids are added onto the chain.
constitutive gene expression
constantly transcribing and translating. products are always needed.
regulated gene expression
product only needed at certain times. expression involves a control mechanism.
deoxyribonuclease
“DNase”, breaks bond between deoxyribonucleotides
14C-amino acid vs 12C-amino acid
12C is a normal amino acid, 14C is a radiolabeled amino acid, and radiation counts can be taken to determine if they are incorporated into proteins.
supernatant
soluble molecules from the cytoplasm
activator (protein)
transcription-level control of gene expression. activator proteins bind to activator binding sites and encourage the binding of RNA polymerase.
activator binding site
sites on DNA near the promoter region where activator proteins bind to DNA strands to encourage RNA polymerase binding.
allolactose
inducer in the lac operon, changing the shape of the lac repressor protein such that it no longer represses lac operon expression.
argCBH operon
operon that produces arginine when its concentration is low. otherwise arginine represses the argCBH operon by binding to arginine repressor protein, preventing its unnecessary synthesis.
arginine
amino acid acting as an inhibitor and product of the argCBH operon.
cAMP receptor protein (CRP)
Transcriptional activator in the presence of cAMP. Previously believed to be involved in the lac operon, but has since been debunked in favor of the EllA inhibitory pathway.
co-repressor
signal molecule that enables a repressor protein in order to inhibit gene transcription.
cyclic AMP (cAMP)
signal molecule for CRP (cAMP receptor protein) that represses gene transcription.
inducer
signal molecule that turns on an activator protein or turns off a repressor protein, to promote gene transcription.
induction
process by which an inducer activates gene transcription, leading to the eventual production of proteins.
lac operon (lacZYA)
encodes for beta-galactosidase (cleaves lactose into glucose and galactose), lactose symporter (brings lactose into the cell), and transacetylase.
lac repressor
repressor that inhibits lacZYA transcription until allolactose is present. allolactose alters the shape of the lac repressor so that it no longer inhibits its expression.
lactose permease
symporter that simultaneously brings lactose and protons into the cell.
MalT
maltose transcriptional activator protein. encourages the transcription of maltose transporter proteins (MalEFG operon) in the presence of maltose. Maltose acts as an inducer, activating the MalT activator protein.
maltose
sugar acting as an inducer towards the MalT maltose activator protein, thus encouraging the expression of maltose transporter protein genes.
maltose operon (malEFG)
operon regulated by MalT that encodes for Maltose transporter proteins.
maltose transporter (Mal E, F, G, K)
expression is activated in the presence of Maltose, binding to the MalT activator protein.
operator
repressor binding site on DNA waiting to be transcribed.
operon vs regulon
an operon is a set of genes cotranscribed by 1 mRNA.
a regulon is a set of genes regulated by the same conditions.
repression
process of reducing gene expression
repressor (protein)
proteins that reduce gene expression by binding to operators in DNA sequences and discouraging the binding of RNA polymerase
B-galactosidase
enzyme that cleaves lactose into glucose and galactose.
transcriptional regulator (protein)
general term for either activator proteins or repressor proteins, working to either encourage or prevent gene expression at the transcriptional level.
CheA
invovled in chemotaxis control mechanism. CheA is phosphorylated first, and phosphorylates CheY next.
CheY
Is phosphorylated by CheA. In this phosphorylated state, encourages clockwise motion of flagellum (tumble)
When CheZ is present and dephosphorylates CheY-P, CheY (unphosphorylated) encourages counterclockwise motion (run).
CheZ
phosphatase enzyme that removes a phosphate from CheY, leading to fluctuations of run and tumble from flagellar bacteria.
EllA subunit of glucose transporter
form of post-translational regulation. the EllA subunit on the glucose transporter inhibits lac permease (when EllA does not have a phosphate attached, as when it is given to glucose for transport into the cell), preventing lactose from being uptaken into the cell. If phosphorylated, EllA no longer inhibits lac permease, and lactose can flow into the cell.
EnvZ
a plasma membrane protein that changes shape in high osmolarity environments to activate kinase (phosphorylation) activity. Phosphorylates OmpR activator protein, which activates transcription of OmpC and micF.
micF
small RNA sequence transcribed by activated OmpR activator protein (high osmolarity environments) that blocks translation of the OmpF gene by competitively blocking the shine dalgarno sequence and start codon.
OmpC
membrane transporter protein that is ideal for high osmolarity conditions. translated when EnvZ phosphorylates OmpR activator protein, meanwhile OmpF is inhibited.
OmpF
membrane transporter protein that is ideal for low osmolarity conditions. translated when EnvZ is inactive. otherwise, EnvZ activation activates OmpR activator protein, translating micF RNA sequence that competitively inhibits translation of OmpF by covering the shine-dalgarno sequence and start codon.
OmpR
regulator protein activated by EnvZ in the OmpR phosphorylation regulation pathway for osmolarity adaptation.
Repellent/attractant receptor
MCPs work as transmembrane receptors that detect environmental attractants or repellents, setting of the chemotaxis response pathways. CheW → CheY-P → CW tumble. or CheW → CheY → CCW run
sensor kinase
protein that senses certain conditions and phosphorylates something under those active conditions
DNA gyrase
2nd protein in DNA replication, unwinds DNA supercoils before the replicating site
DNA helicase
3rd protein in DNA replication, uses ATP to unwind DNA double helix
DNA replication
process of copying DNA for daughter cells before cell division.
filamentous temperature sensitive (fts) mutant (ftsZ)
special mutated cells that form long filaments under high temperature conditions. in these mutant cells FtsZ proteins that usually pinch the cleft to divide cells do not function properly at high temperature, leading to filamentous cells
genotype
genetic makeup.
mutant
has an inherited change in the nucleotide sequence of the microbial genome.
origin binding protein
1st protein in DNA replication. Binds to origin of replication.
origin of replication
DNA sequence attracting an origin binding protein, signaling for DNA polymerase to bind later.
phenotype
physical, observable attributes of an organism. impacted by genotype and environmental conditions.
RNA primase
5th protein of DNA replication. Adds RNA nucleotides with a 3’ OH group, allowing DNA polymerase to work. This protein is associated with helicase, and acts as the protein unwinds the double helix.
screen
mutant phenotype does not lead to conditions under which mutant phenotype cannot grow. think velvet imprint of desired colonies as opposed to growth on antibiotic plate (selection).
selection
mutant phenotype allows growth under conditions in which wild type cannot grow.
single-strand binding protein
4th protein in DNA replication. holds the single strands of DNA apart so they can be replicated, accessed by DNA polymerases.
wild type
“normal” strain
3’ to 5’ exonuclease
is able to cleave phosphodiester bonds in the 3’ to 5’ direction. allows for proofreading activity. opposite of direction of replication (5’ to 3’). both DNA poly III and DNA poly I have 3’ to 5’ exonuclease activity.
5’ to 3’ exonuclease
is able to cleave phosphodiester bonds in the 5’ to 3’ direction. allows for the breakdown and subsequent replacement of RNA primers written in 5’ to 3’ manner.
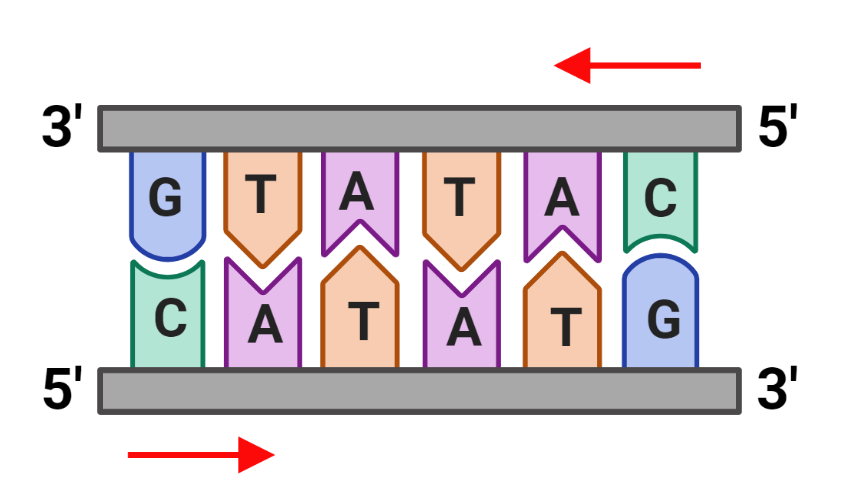
antiparallel
parallel, but oriented in the opposite direction
3’ — 5’
5’ — 3’
deletion
mutation that removes nucleotides from a sequence.