MCB*2050: Post-Midterm Content
1/428
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
429 Terms
traditional cell biology
“The multidisciplinary study of the composition, structure, and function of the various subcellular organelles and other cellular components in evolutionarily diverse organisms.”
cell biology today
“The post-genomics, cellular imaging revolution”
today’s problem for cell biology
~25-50% of genes encode proteins of unknown function(s).
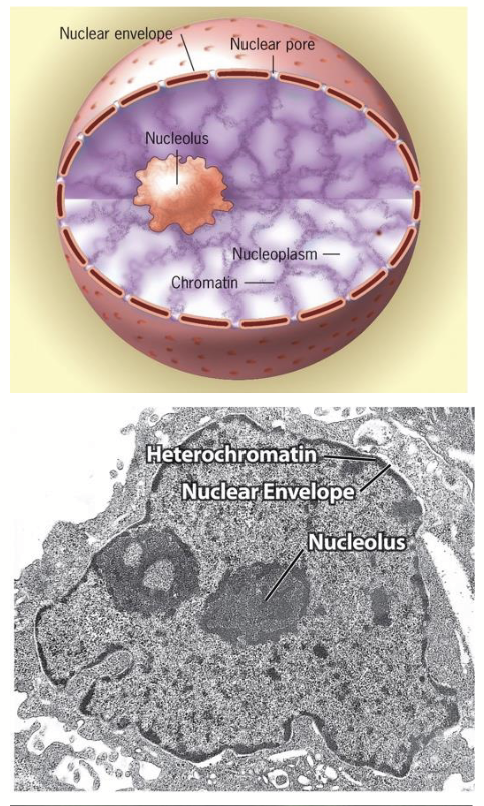
nucleus
Largest organelle; typically one per cell.
Size varies from cell-to-cell and between organisms.
Usually determined by cell size (i.e. cytoplasmic volume)
Increases during development and in cancer cells - used for cancer diagnosis/prognosis
Primary difference between prokaryotes and eukaryotes:
Eukaryotes: membrane-bound
Prokaryotes: “region” where chromosome is located; less DNA, less DNA packaging, and limited/no RNA processing
nucleoid
In prokaryotes, the “region” where the chromosome is located. Contains less DNA, less DNA packaging, and limited/no RNA processing.
eukaryotic cell
Larger, structurally and functionally more complex interiors. Possess single and double membrane-bound organelles.
Cellular “compartmentalization” allows for larger size and segregation and organization of specific cellular functions.
Each organelle contains both unique and common factors for functioning (e.g., metabolism) and their biogenesis (formation), maintenance (+ life cycle), and turnover.
nucleus function
Responsible for two main functions:
Compartmentalization of the cellular genome and its activities.
e.g. site of DNA replication, transcription, and RNA processing
e.g. site where translation components (ribosomes, mRNAs, tRNAs) are synthesized
Coordination of cellular activities.
e.g. control of metabolism, protein synthesis, reproduction (cell division), etc.
nucleus: gene expression
Separation of the cytoplasm from the genome allows for unique spatial & temporal regulation of gene expression in eukaryotes.
Prokaryotes: mRNAs translated while transcription is in progress.
Eukaryotes: mRNAs undergo post-transcriptional processing (e.g., splicing) before transport out of nucleus and then translated in cytoplasm or at ER
Additional control of gene expression in eukaryotes: nucleus (nuclear envelope) limits access of transcription factors (proteins) from cytoplasm to genome.
All because of compartmentalization!
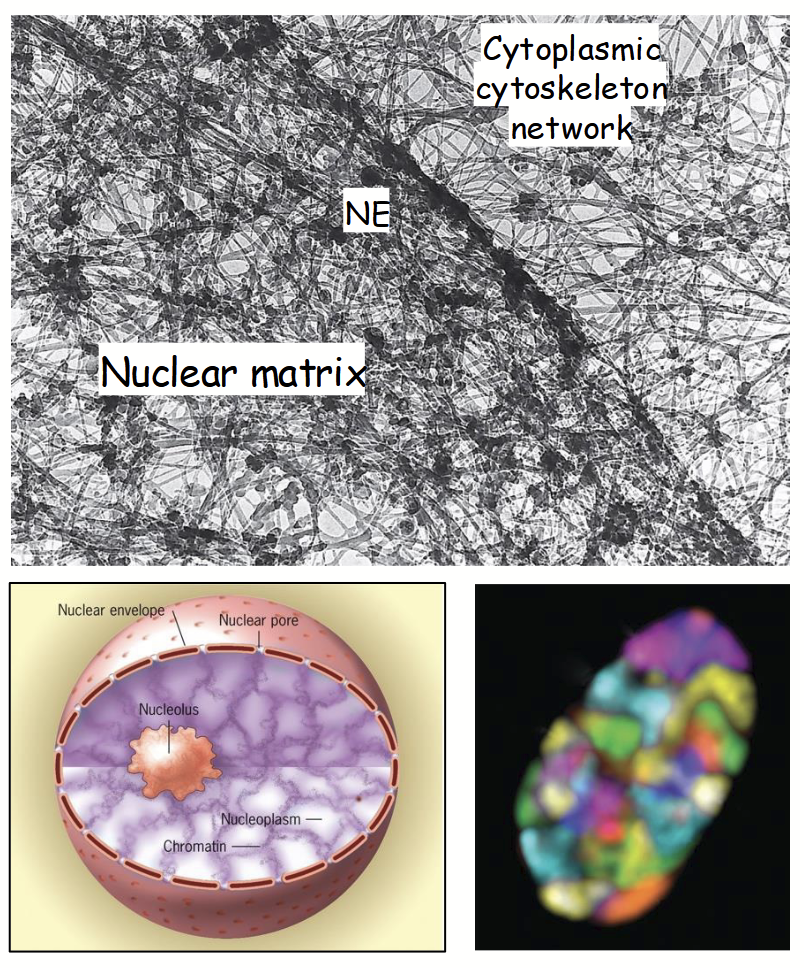
nucleoplasm
Nuclear structure: internal organization
Fluid-filled interior of nucleus - highly organized. Consists of >30 specialized regions (subdomains) that participate in specific functions (note: they are not membrane-bound).
subdomains: e.g. nucleolus, chromosomes during interphase, nuclear speckles
nucleolus
The most conspicuous nuclear subdomain; irregularly shaped, dense, and granular in appearance.
Size and number (1-5) depend on metabolic activity of cell (↑cellular activity, ↑protein synthesis, ↑size/number).
Function in producing ribosomes:
Site of ribosomal DNA (rDNA) gene transcription, rRNA processing, and initial stages of ribosomal subunit (rRNA + protein) assembly.
Final assembly of ribosomes (used for protein synthesis) occurs in cytoplasm.
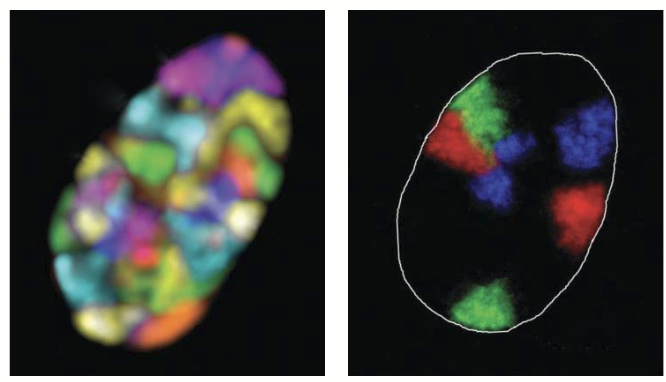
chromosomes
During interphase, are organized into discrete subdomains within the nucleus.
Location of a gene is often related to its activity.
Most actively transcribed genes (i.e. decondensed chromatin) are found at periphery of chromosomal “subdomains”.
interchromosomal channels
Regions between chromosome subdomains that serve as barriers to prevent unwanted DNA-DNA and/or DNA-protein interactions.
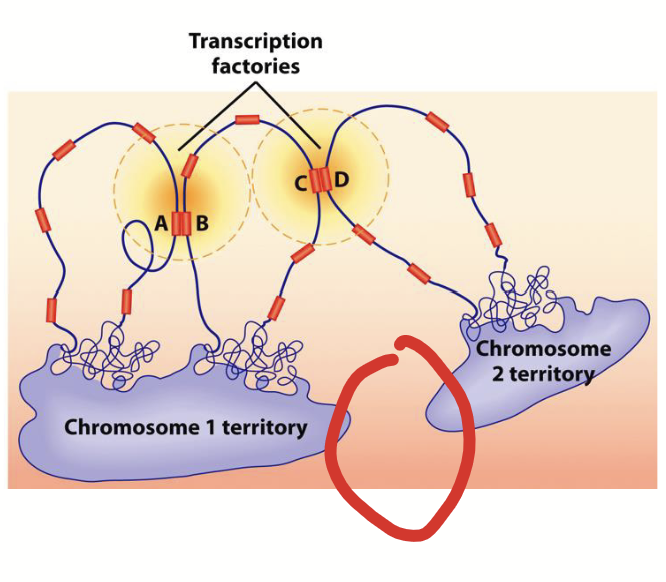
chromosomal subdomains
Active genes (euchromatin) from different subdomains (or from different regions of same chromosome) extend into interchromosomal channels to form transcription factories where transcription factors are concentrated.
transcription factories
Formed when active genes (euchromatin) from different subdomains (or from different regions of same chromosome) extend into interchromosomal channels. Where transcription factors are concentrated.

interchromosomal interactions
“Kissing chromosomes”.
Gene regulatory regions from one chromosome activate gene(s) on another chromosome. The chromosomes physically interact.
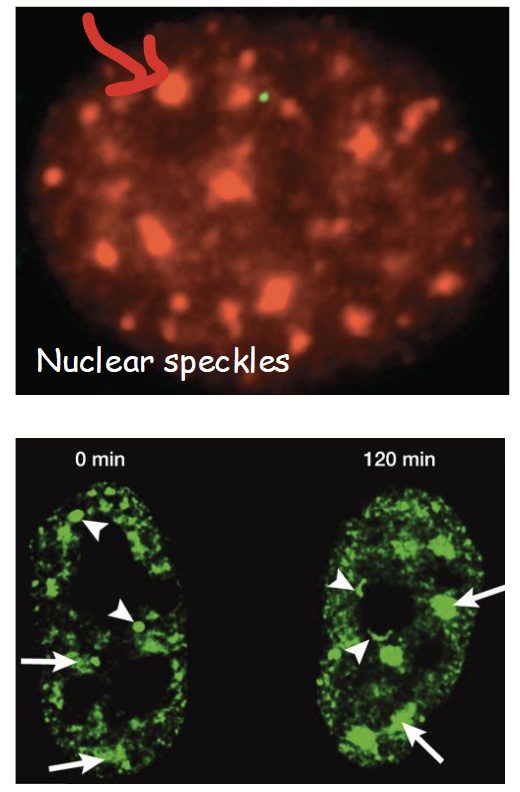
nuclear speckles
Nuclear subdomains where mRNA splicing factors are concentrated (i.e. where pre-mRNA processing occurs).
Often located in interchromosomal channels next to transcription factories.
Numerous and highly dynamic - often move quickly and grow/shrink and change in number depending on needs of cell.
Grows when there is more mRNA (because more splicing required).
nucleus
Nuclear structure: internal organization
Insoluble fibrillar-like protein network (“mesh”) distributed throughout nucleoplasm.
Framework that provides support + surface for movement, like a highway.
Analogus to cytoskeleton network in cytoplasm.
Composed of 3 major filament systems:
Microfilaments
Microtubules
Intermediate filaments
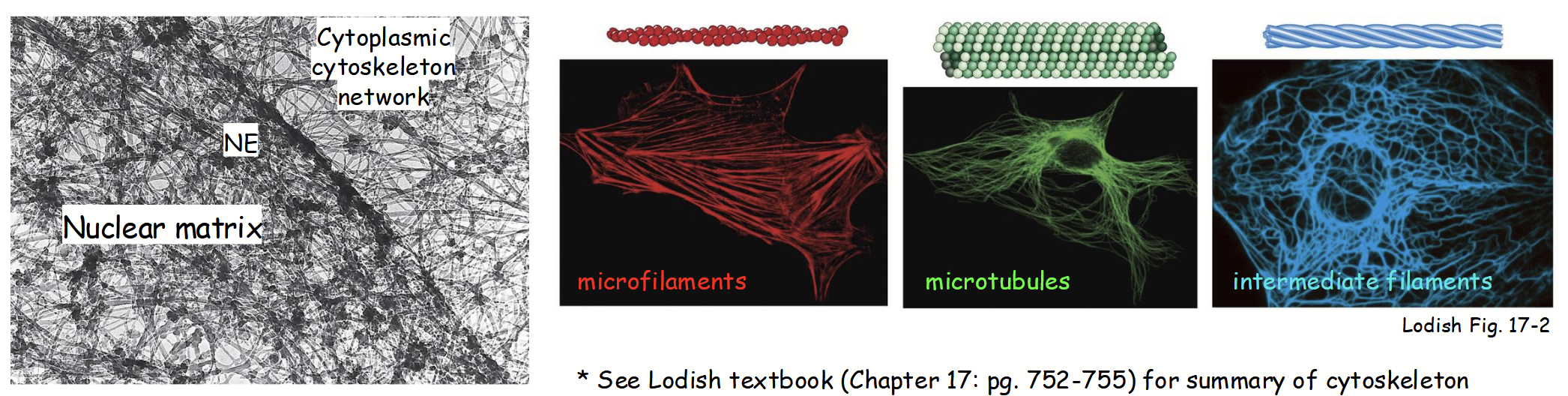
nuclear matrix
Nuclear structure: internal organization
Serves a structural role - maintains overall shape of nucleus.
Serves as a “scaffold” - responsible for organization nuclear subdomains and anchoring protein factors (e.g. proteins involved in DNA replication, transcription, RNA processing, etc.).
Very little is known about composition and assembly/disassembly.
nuclear envelope
Nuclear structure: internal organization
Separates the contents (e.g. genome) of nucleus from the surrounding cytoplasm.
Serves as a barrier - required regulated passage of molecules (e.g. RNA, proteins) between nucleus and cytoplasm.
Establishes unique composition of nucleus (compared to cytoplasm) and spatially regulated gene expression.
Provides structural framework for nucleus.
Composed of three main parts:
Nuclear membranes
Nuclear lamina
Nuclear pore complexes
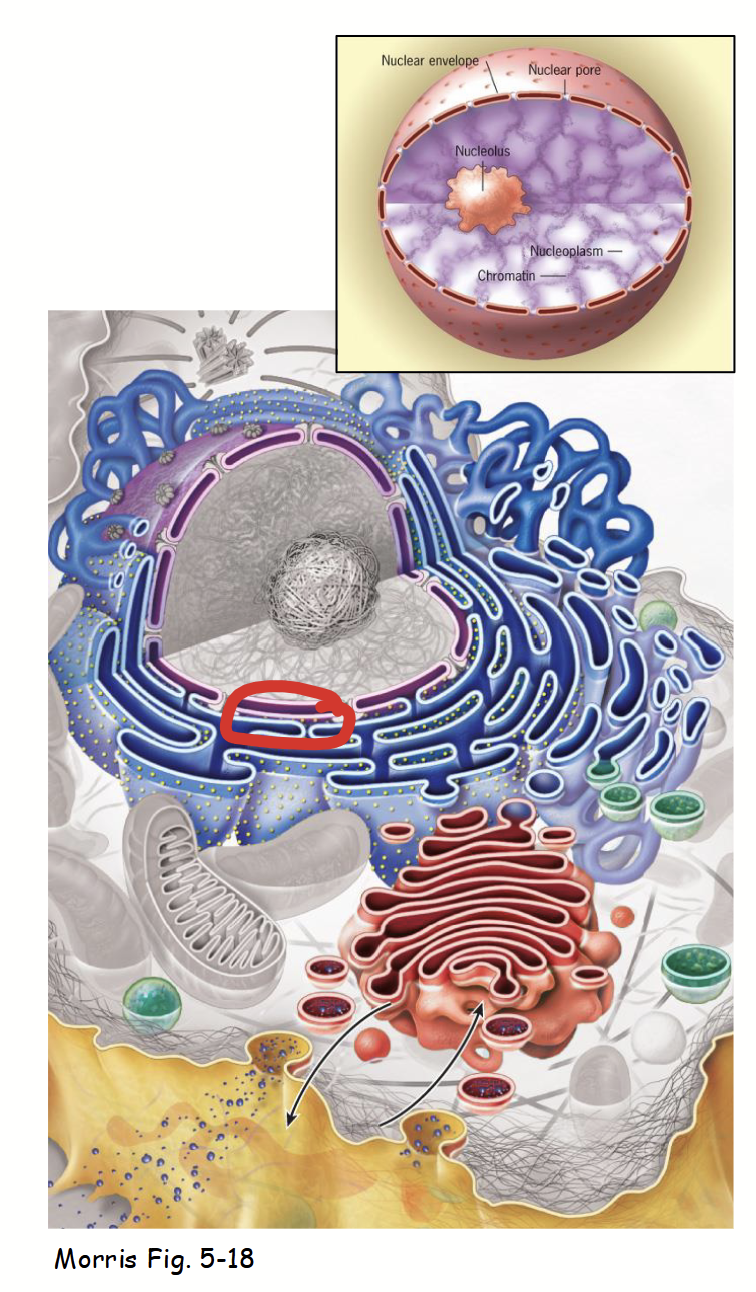
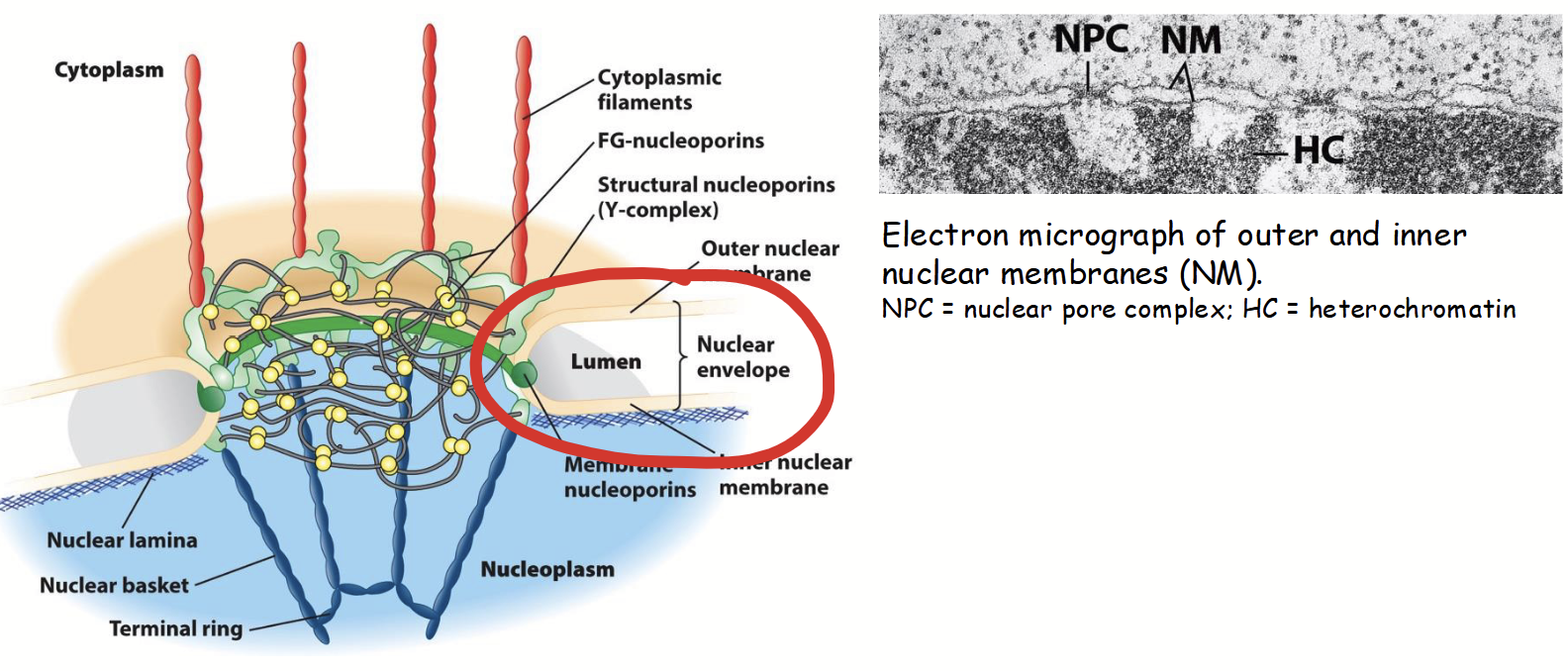
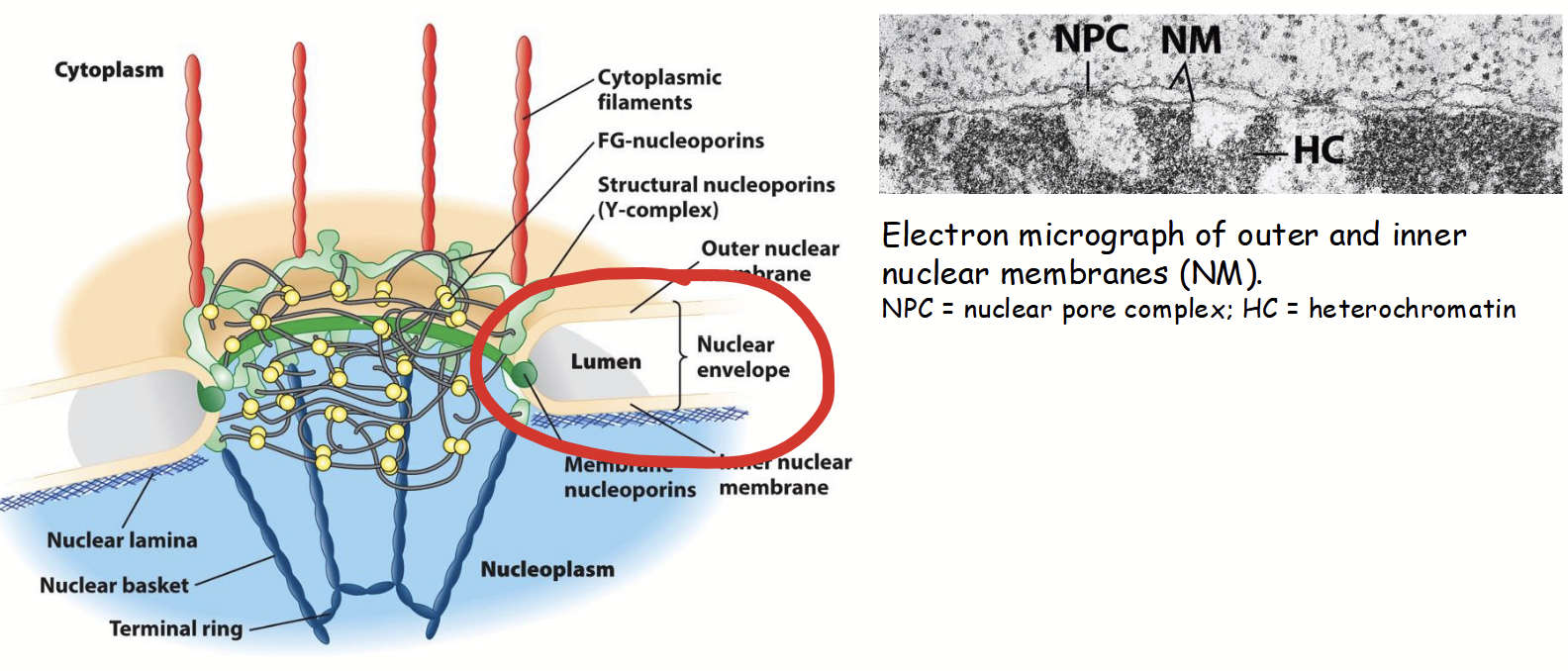
nuclear membranes
Part of the nuclear envelope.
Inner and outer: two concentric membranes (phospholipid bilayers) arranged in parallel.
Inner and outer are separated by nuclear envelope lumen (10-50 nm diameter; usually very consistent).
Membranes serve as barriers to passage of ions, solutes, and macromolecules between the nucleus and the cytoplasm.
inner and outer nuclear membranes
Two concentric membranes (phospholipid bilayers) arranged in parallel. Part of the nuclear envelope.
Separated by its lumen.
Joined (highly surved) at nuclear pore complexes/

nuclear envelope lumen
Separate the inner and outer nuclear membranes. 10-50 nm in diameter, usually very consistent.
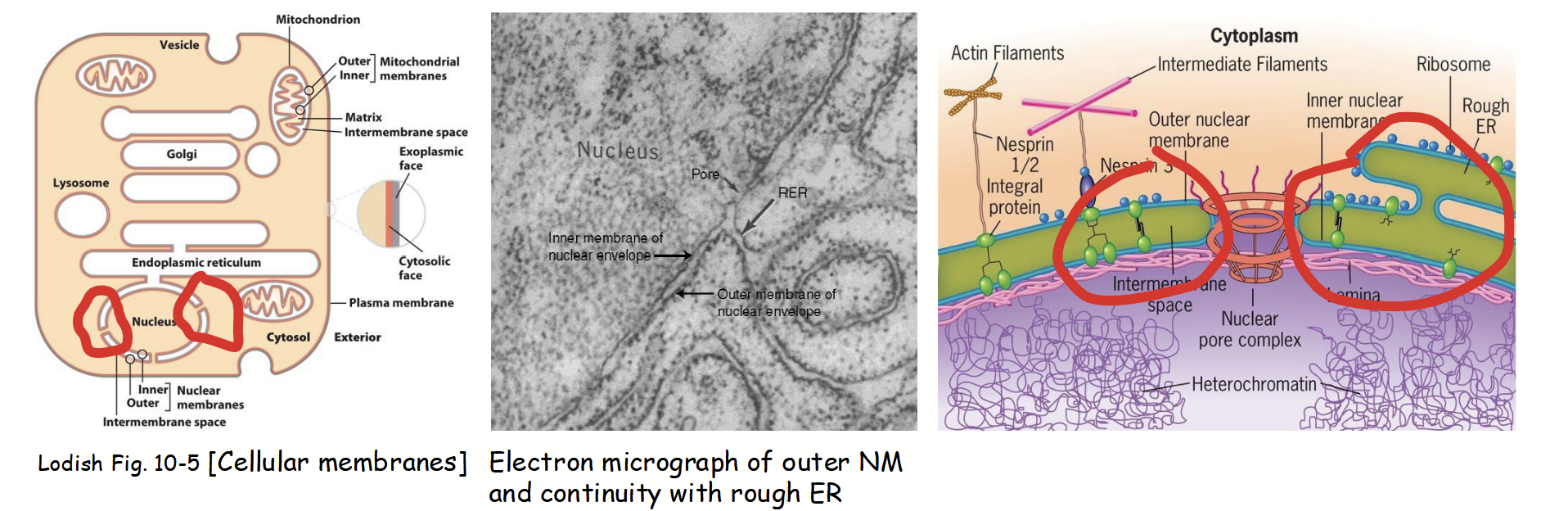
outer nuclear membrane
Part of the nuclear membranes that is continuous with the rough endoplasmic reticulum (RER).
Ribosomes attached to cytoplasmic surface (functionally similar to RER).
Nuclear envelope envelope lumen is continuous with ER lumen.
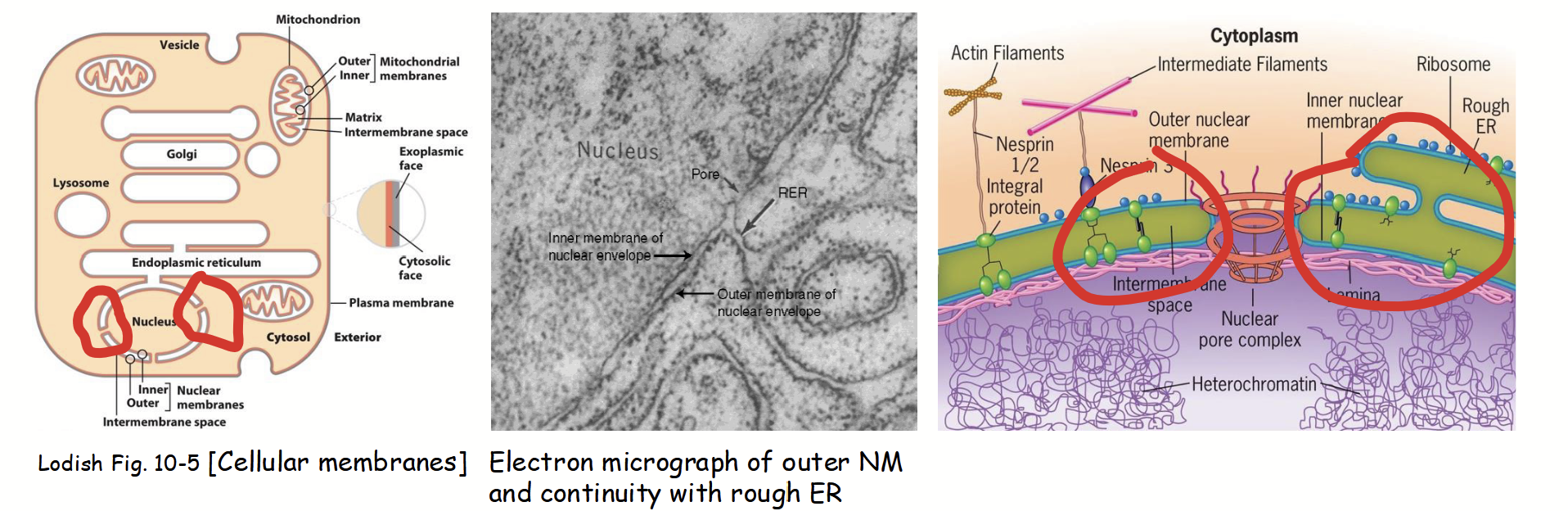
inner nuclear membrane
Part of the nuclear membranes with a unique protein composition (functionally distinct from the outer membrane).
nuclear lamina
Part of the nuclear membranes.
Located on inner surface (i.e. nucleoplasmic side) of nuclear inner membrane.
Network (“mesh”) of long, filament-like proteins.
ABC nuclear lamins: evolutionarily related to proteins that form intermediate filaments in cytoskeleton network.
Provides mechanical support to nuclear envelope (binds to nuclear inner membrane integral proteins).
Serves as scaffold for attachment of chromatin and nuclear matrix to nuclear envelope.
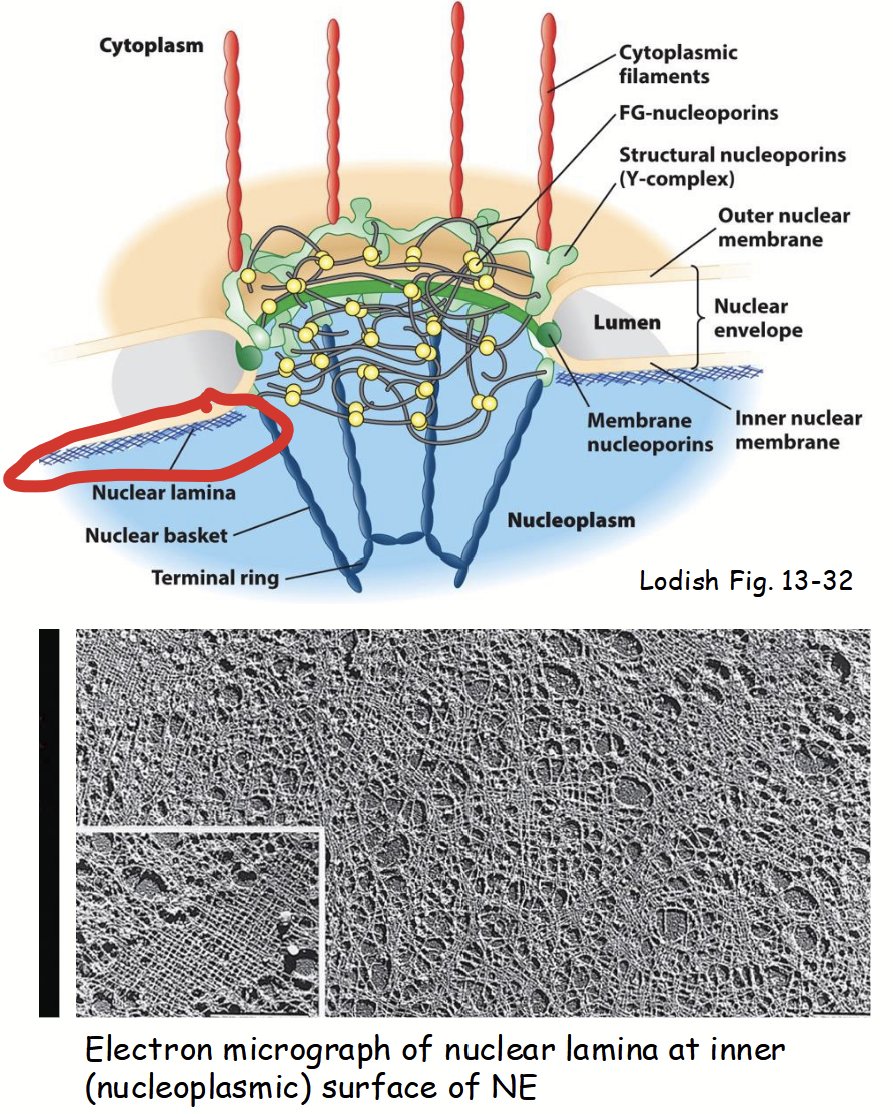
ABC nuclear lamins
Nuclear lamina evolutionarily related to proteins that form intermediate filaments in cytoskeleton network.
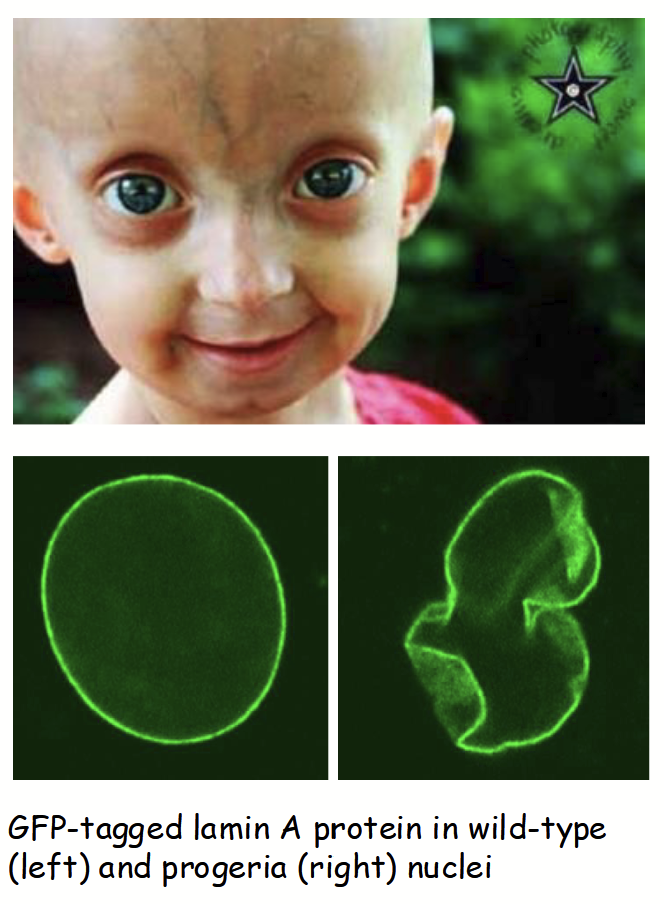
nuclear lamina mutations
Mutations in LAMIN genes responsible for several human diseases.
e.g. Hutchinson-Gilford Progeria Syndrome
Rare, characterized by premature aging in children (e.g. hair loss, wrinkles, artery damages) - death by early adolescence
Due to a point mutation (sproadic - occurs in embryo development) in LAMIN A gene leading to truncated lamin protein
Results in destabilization/breakdown of nuclear lamina, causes aberrant changes in nuclear (envelope) morphology and function
Recently: promising advances using CRISPR/Cas9 genome editing-based (gene) therapy in mice
nuclear pore complex (NPC)
Part of the nuclear membranes.
Channels (“doorways”) in nuclear envelope.
Responsible for regulated trafficking (import and export) of all substances between nucleus and cytoplasm.
Small, polar molecules (e.g. nucleotides for DNA/RNA synthesis)
RNAs: mRNA, tRNA, rRNA
Proteins (e.g. transcription factors, RNA-binding proteins, ribosomal (subunit) proteins, and cyclins)
Typically, 3000-4000 per nucleus; number related to nuclear activity.

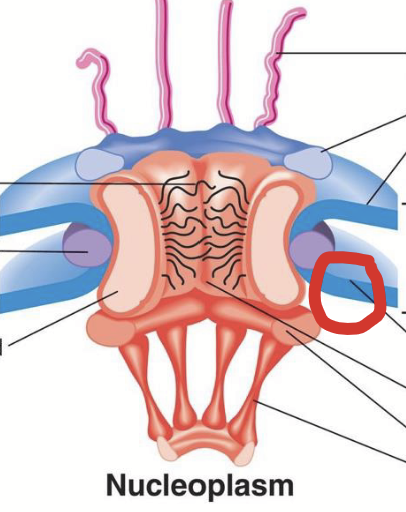
nuclear pore complex (NPC) structure
Large, highly complex structure (~30X > ribosome).
Composed of ~40 different proteins called nucleoporins (“Nups”).
Overall structure: 8-fold symmetrical structure organized around large, central aqueous channel.
Consists of several parts:
Central scaffold
FG nucleoporins.
Y-complexes
Cytoplasmic filaments
Nuclear basket
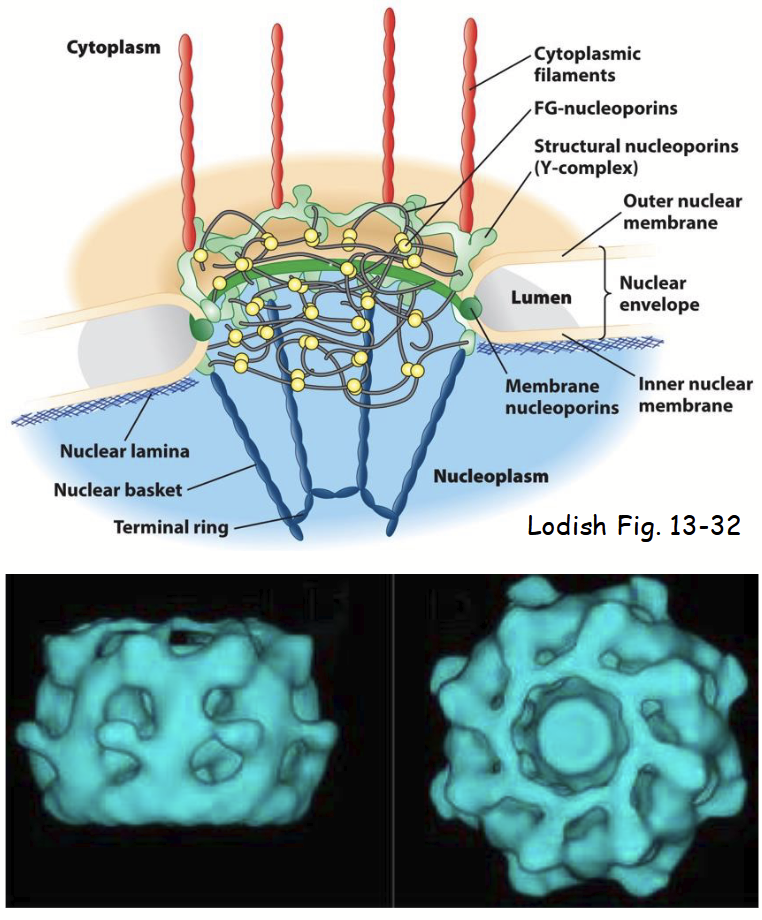
nucleoporins (Nups)
~40 different proteins that make up the nuclear pore complex (NPC).
Highly conserved among all eukaryotes.
Include both integral and peripheral inner and outer nuclear membrane proteins.
Several related to COPII proteins involved in vesicle formation at ER.
Common evolutionary origin: both these and COPII proteins function to deform (curve) membranes.
central scaffold
Part of the nuclear pore complex (NPC).
Composed of integral/trans membrane-bound nucleoporins.
Anchors NPC to nuclear envelope membranes (at junction of outer and inner membranes).
Forms aqueous central channel (~20-40 nm wide pore).
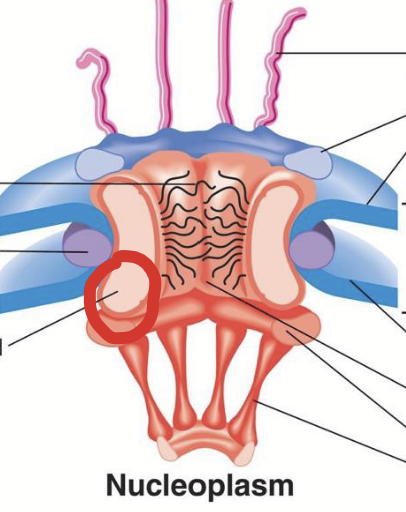
central channel
Part of the nuclear pore complex (NPC).
Aqueous channel formed by the central scaffold in the nuclear pore complex. A ~20-40 nm wide pore.
The inner surface is lined by FG nucleoporins.
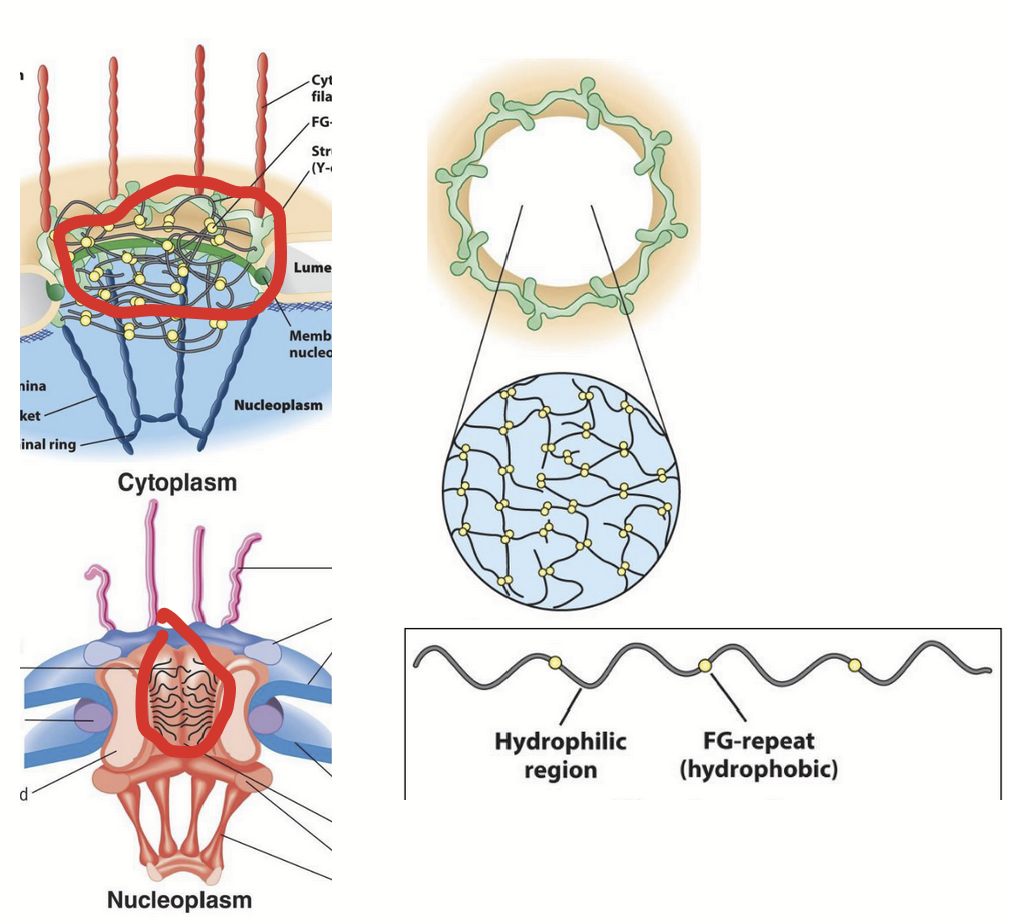
FG nucleoporins
Part of the nuclear pore complex (NPC).
“Filament-like” nucleoporins that line the inner surface of the central channel.
Possess unique amino acid composition.
Hydrophilic polypeptides with short repeats of hydrophobic domains enriched in Phe and Gly.
Possess a unique, highly disordered secondary structure.
Extended/flexible organization fills central channel.
Domains extend into central channel.
Form “mesh” (sieve-like gel) that limits diffusion of macromolecules larger than ~40 kDa (~39 nm diameter).
Small molecules move freely through NPC in either direction (e.g. nucleotides - DNA and RNA synthesis).
Molecules >40 kDa unable to pass through NPC freely.
e.g. RNA and most proteins must be selectively imported/exported by active process (active process).
Size-exclusion limit for NPC.
Based on studies using microinjected gold particles of varying sizes and coated with nuclear protein.
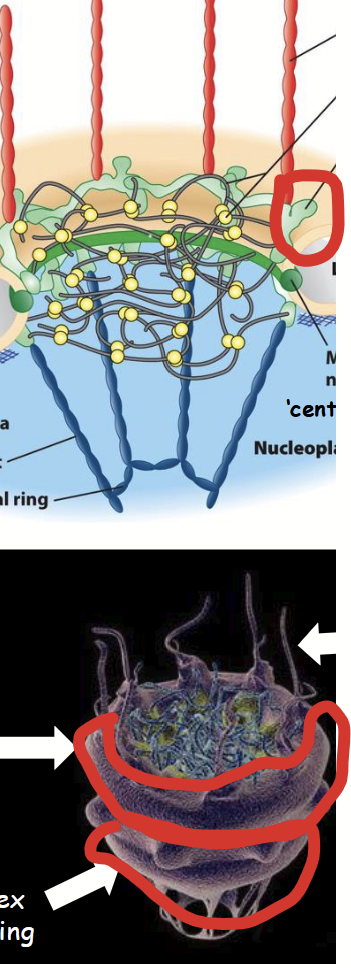
Y-complexes
Part of the nuclear pore complex (NPC).
Includes cytoplasmic ring and nuclear rings; both composed of structural nucleoporins.
Located on cytoplasmic and nuclear (nucleoplasmic) side of NPC, respectively.
Linked to central scaffold and also to cytoplasmic filaments or nuclear basket.
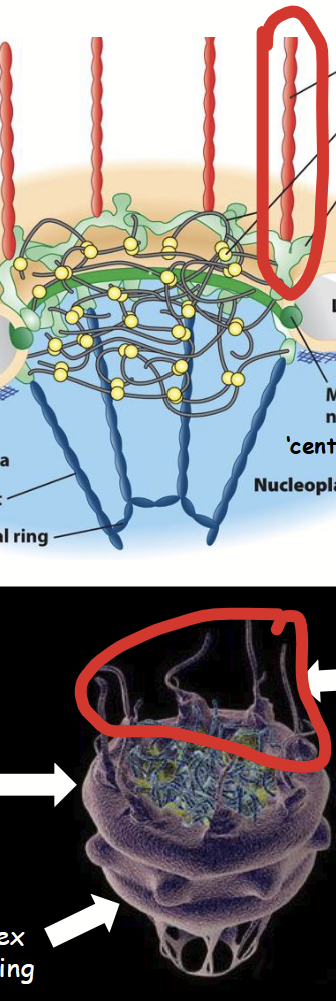
cytoplasmic filaments
Part of the nuclear pore complex (NPC).
Long, filament-shaped (structural) nucleoporins that extend into the cytoplasm.
Involved in nuclear receptor-cargo protein recognition and import from cytoplasm.
nuclear basket
Part of the nuclear pore complex (NPC).
“Basket-like” structure (also made of structural nucleoporins) located on nuclear (nucleoplasmic) side of NPC.
Linked to Y-complex nuclear ring.
Involved in nuclear receptor-cargo protein important and export to cytoplasm (both forward AND reverse processes)
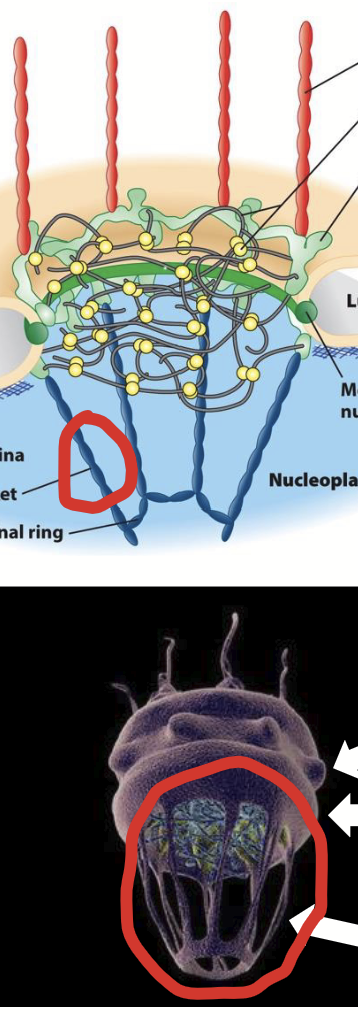
nucleocytoplasmic transport via NPC
Among most congested bi-directional trafficking pathways in cell.
Includes variety of cytoplasm-to-nucleus (import) and nucleus-to-cytoplasm (export) trafficking pathways.
Involves wide range of “cargo”.
All proteins required for DNA replication, transcription, splicing, ribosome assembly, chromatin packing (histones), nuclear matrix proteins, lamins, etc., are imported into the nucleus from the cytoplasm.
All RNA (mRNA, tRNA, rRNA), partially assembled ribosomes (required for protein synthesis in cytoplasm), and some proteins are exported out of the nucleus into cytoplasm.
Molecular mechanism is well understood: requires energy, specific protein receptors, and unique targeting signals.
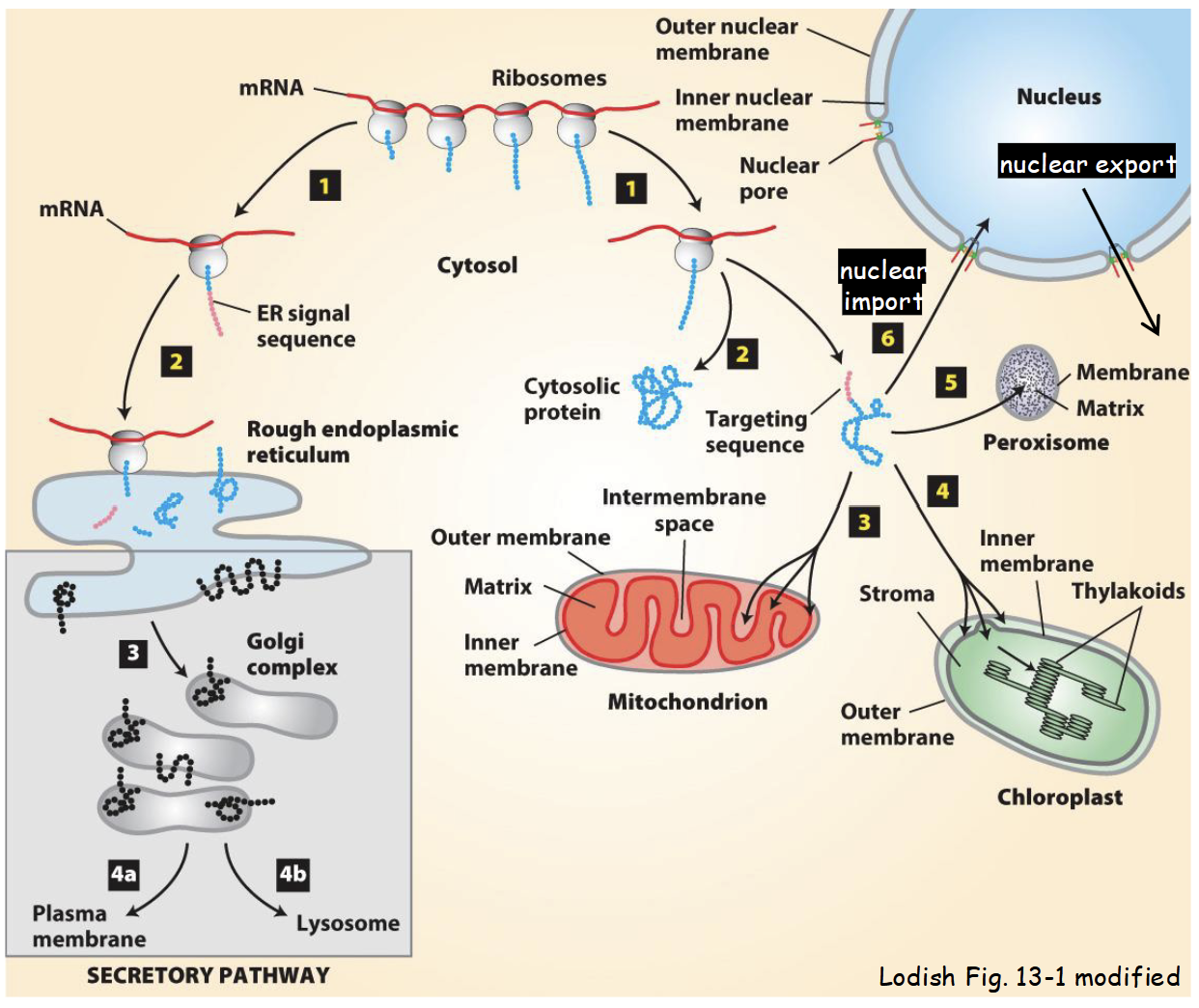
cytoplasm-to-nuclear transport
Most nuclear-imported proteins (e.g. transcription factors, nuclear matrix proteins, histones, lamins, cyclins, etc) contain a nuclear localization signal (NLS).
Characterization of different NLSs led to identification of factors necessary for nuclear import of proteins from cytoplasm.
Including transport receptors: mobile proteins responsible for moving (“ferrying”) protein “cargo” across nuclear envelope.
Karyoferins: large family of receptor proteins responsible for moving macromolecules either into nucleus (importins) or out of nucleus (exportins).
Protein import is a multi-step process with five main steps.
nuclear localization signal (NLS)
Specific stretch/sequence of amino acids recognized by nuclear receptor proteins. Serves as the “zipcode” to mediate targeting of protein from cytoplasm to nucleus.
Involved in cytoplasm-to-nuclear transport.
Proteins can have more than one + more than one nuclear export signal (NES).
Identified in proteins based on mutational analyses.
An amino acid sequence that is both necessary and sufficient for cytoplasm-to-nuclear targeting.
Necessary: if sequence (or a portion thereof) is mutated, then modified protein fails to target to nucleus (i.e. mutant protein is mislocalized to cytoplasm).
Sufficient: if sequence linking to non-nuclear (“passenger”) protein is capable of redirecting resulting fusion protein to nucleus.
Several different types: based on different (unique) amino acid sequences.
Classic
Bipartite
classic NLS
Most common NLS. Consists of a short stretch of positively-charged (basic) amino acid residues.
e.g. KKQRKK - in simian virus 40 large T antigen protein
bipartite NLS
NLS composed of two short stretches of basic amino acids and a 7-10 amino-acid-long “spacer” sequence.
e.g. KR[PAATKAGQA]KKKK - in nucleoplasmin
ARC1
Protein required for plant pollination. Involved in signal-transduction pathway for recognition of self-incompatible pollen at plasma membrane.
Shuffles between nucleus and cytoplasm.
Possesses both an NLS and NES.
Before pollination: NLS > NES results in localization mostly in nucleus.
During (self-pollination): NLS disrupted due to phosphorylation of adjacent AA residue(s).
NLSPhos < NES results in localization mostly in cytoplasm.
In cytoplasm: functions in proteosome-dependent turnover of other proteins - prevents self-pollination.
identification of NLS in ARC1
ARC1 localized to nucleus: contains putative classic NLS (amino acids 261-266) and putative nuclear export signal.
transport receptors
Mobile proteins responsible for moving (“ferrying”) protein “cargo” across nuclear envelope.
karyoferins
Large family of receptor proteins responsible for moving macromolecules either into nucleus (importins) or out of nucleus (exportins).
= nucleus
“karyo”
cytoplasm-to-nucleus transport: step 1
Nascent (newly-synthesized) NLS-containing “cargo” protein is recognized in cytoplasm by importin.
Importin: heterodimeric protein.
Consists of two distinct subunits: importin α and importin β.
Importin α subunit recognizes and binds to basic residues in “cargo” protein’s NLS.
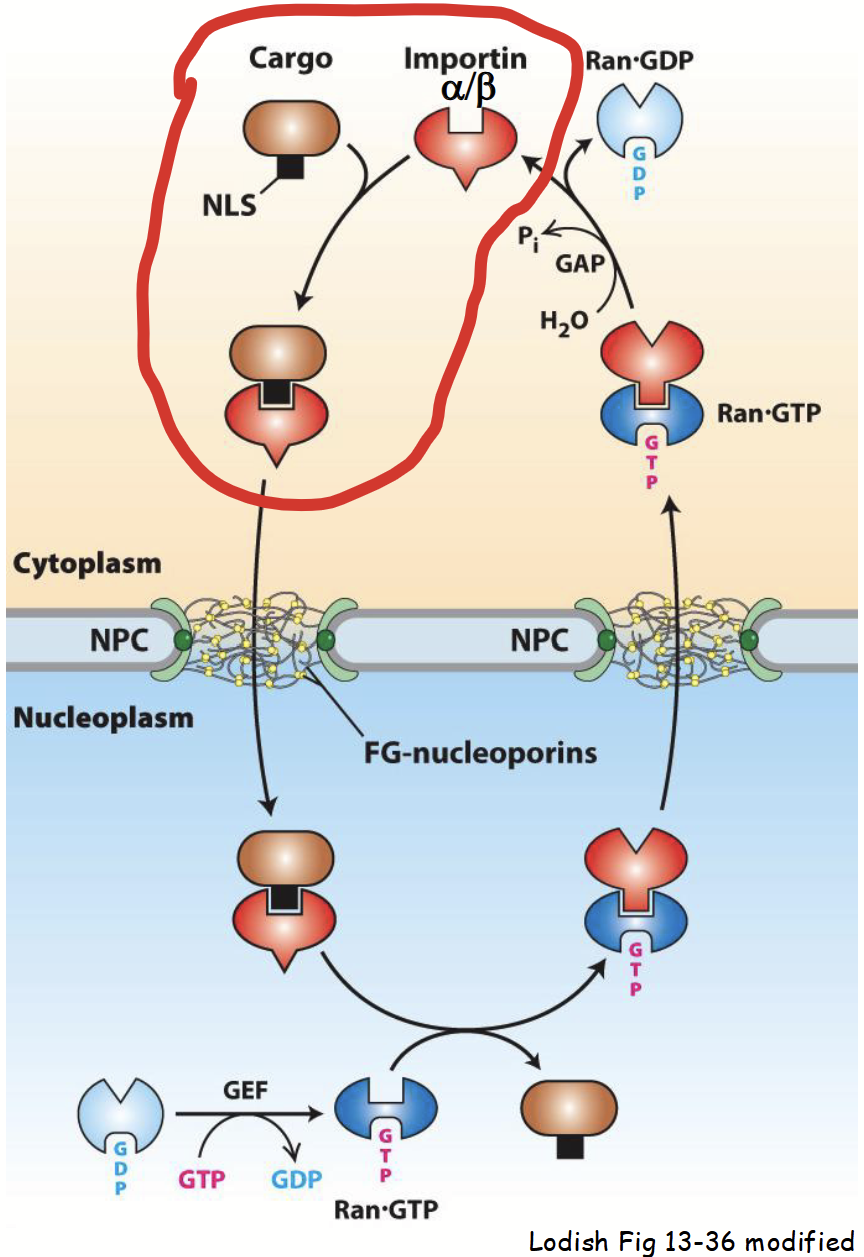
importin
Heterodimeric karyoferin made up of an α subunit and a β subunit. Takes part in cytoplasm-to-nucleus transport.
cytoplasm-to-nucleus transport: step 2
“Cargo” protein-importin receptor complex moves through cytoplasm, towards nucleus (via importin’s ability to bind cytoskeleton).
Cytoskeleton elements serves as “highways” for almost all types of intracellular transport (e.g. movement of proteins, RNA, organelles, etc.).
At the surface of the nucleus…
Importin β subunit of cargo protin-importin receptor complex binds to cytoplasmic filaments at NPC.
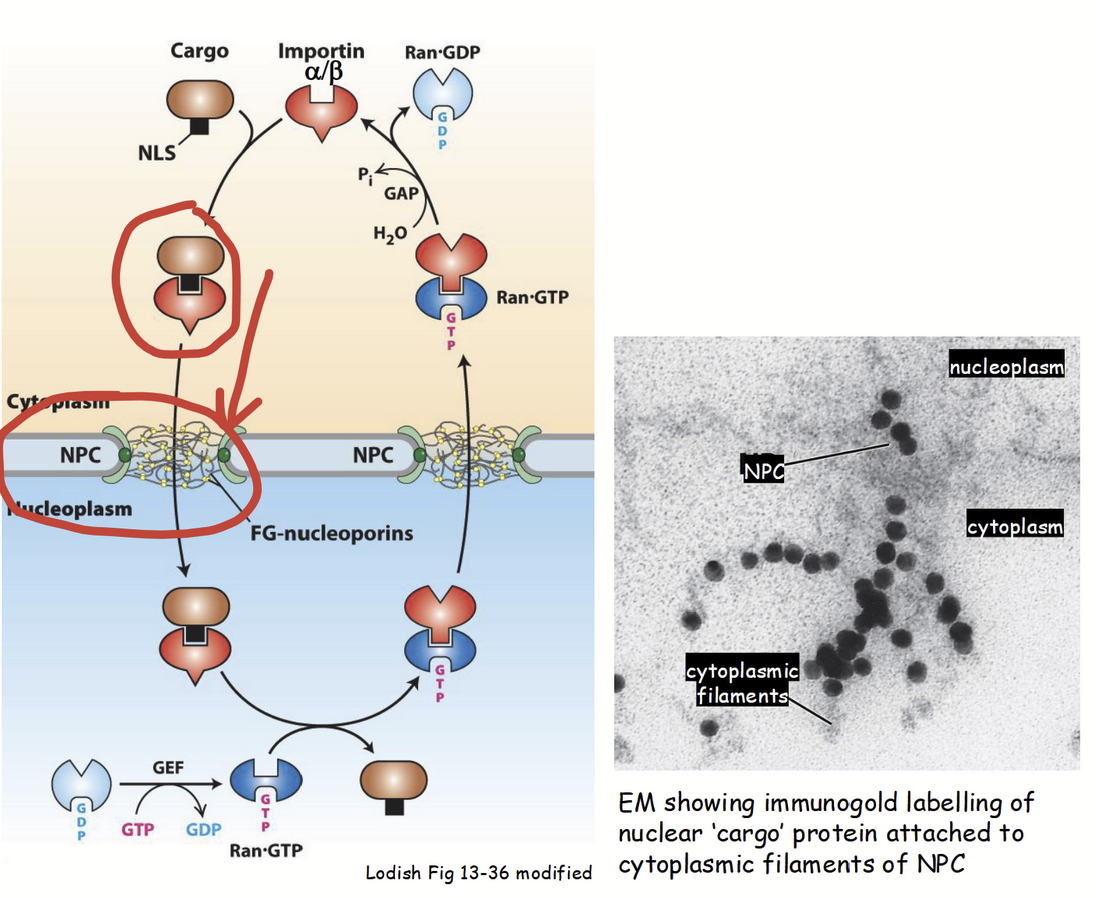
cytoplasm-to-nucleus transport: step 3
“Cargo” protein-importin receptor complex is translocated through central channel of NPC.
Translocation process is not well understood.
Current model: cargo-receptor complex successively interacts with hydrophilic and FG domains of FG nucleoporins in central channel.
Interactions “untangle” the FG-domain network and allow for translocation through central channel.
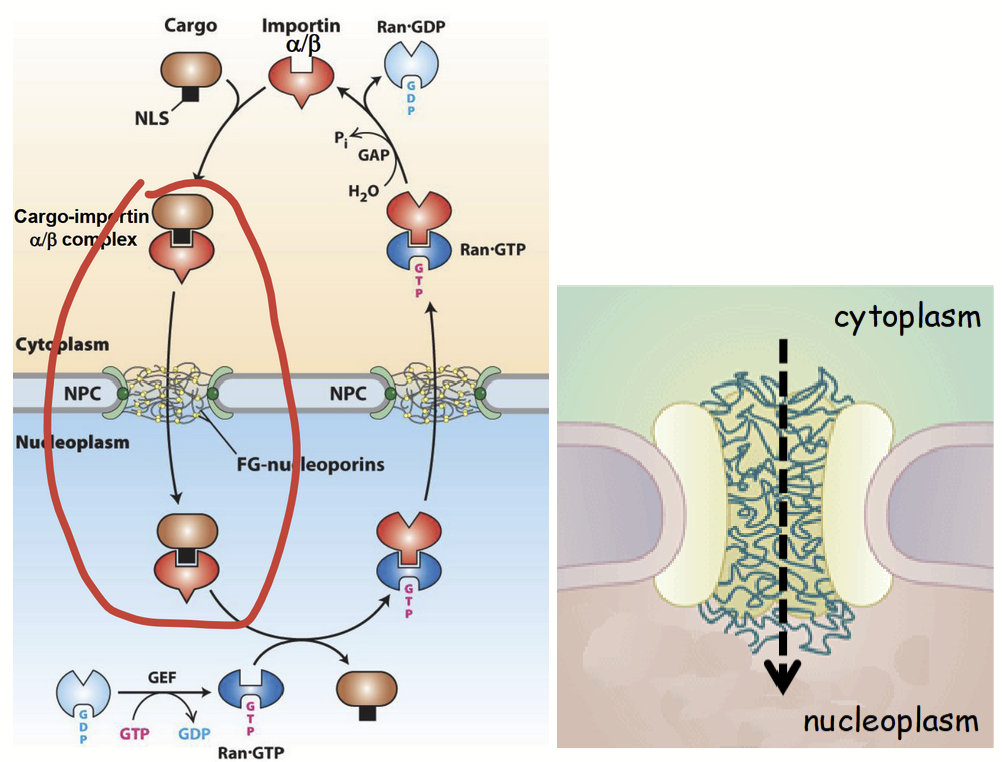
cytoplasm-to-nucleus transport: step 4
“Cargo”-receptor complex associates with nuclear basket on inner surface of NPC.
“Cargo”-receptor complex binds to Ran-GTP (via importin β), resulting in its released from NPC (basket) and disassembly into nucleoplasm.
Import of the NLS-containing “cargo” protein into nucleus is accomplished.
NOTE: NLS is NOT proteolytically cleaved from “cargo” protein, unlike most other organelle targeting signals; allows for re-import of nuclear proteins.

Ran
Small GTPase.
Protein’s conformation and activity regulated by GTP binding and hydrolysis
Exists in two distinct states:
Ran-GTP (active GTP-bound form)
Ran-GDP (inactive GDP-bound form)
A steep concentration gradient of Ran-GTP between nucleus and cytoplasm.
[Ran-GTP]nucleus > [Ran-GTP]cytoplasm
![<ul><li><p>Small <strong>GTPase</strong>.</p><ul><li><p>Protein’s conformation and activity regulated by GTP binding and hydrolysis</p></li></ul></li><li><p>Exists in two distinct states:</p><ul><li><p><strong>Ran-GTP</strong> (active GTP-bound form)</p></li><li><p><strong>Ran-GDP</strong> (inactive GDP-bound form)</p></li></ul></li><li><p>A steep concentration gradient of Ran-GTP between nucleus and cytoplasm.</p></li><li><p>[Ran-GTP]<sub>nucleus</sub> > [Ran-GTP]<sub>cytoplasm</sub></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/940f6647-035e-4907-a7c6-1a8191672336.png)
GEF
Nuclear protein thar promotes conversion of Ran-GDP to Ran-GTP.
Maintains a high [Ran-GTP] in nucleus (i.e. promotes the GTPase activity of Ran).
GAP
Cytoplasmic protein that promotes hydrolysis of Ran-GTP to Ran-GDP.
Maintains low [Ran-GTP] in cytoplasm.
Ran-GTP gradient
A steep concentration gradient of Ran-GTP between nucleus and cytoplasm.
[Ran-GTP]nucleus > [Ran-GTP]cytoplasm
Determines directionality of nucleocytoplasmic transport.
GTP hydrolysis provides energy required for nucleocytoplasmic transport.
cytoplasm-to-nucleus transport: step 5
Ran-GTP bound importin β subunit moves back to cytoplasm due to [Ran-GTP] gradient.
[Ran-GTP]nucleus > [Ran-GTP]cytoplasm
In cytoplasm, GTP on Ran-GTP is hydrozyled via accessory protein GAP.
Ran-GDP released from importin β.
Importin β used for another round of nuclear protein import.
Fate of importin α in the nucleus? Cargo proteins exported out of nucleus?
![<ul><li><p><strong>Ran-GTP bound importin β subunit</strong> moves back to <u>cytoplasm</u> due to [Ran-GTP] gradient.</p><ul><li><p>[Ran-GTP]<sub>nucleus</sub> > [Ran-GTP]<sub>cytoplasm</sub></p></li></ul></li><li><p>In cytoplasm, GTP on Ran-GTP is hydrozyled via accessory protein GAP.</p><ul><li><p>Ran-GDP released from importin β.</p></li></ul></li><li><p>Importin β used for another round of nuclear protein import.</p></li><li><p><strong><em><u>Fate of importin α in the nucleus? Cargo proteins exported out of nucleus?</u></em></strong></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/81f514f6-88e3-4a71-937c-d9c49ef51516.png)
fate of importin α in the nucleus
How does importin α (transport receptor subunit) move back to the cytoplasm in order to participate in additional rounds of import?
Importin α binds to exportin.
Karyopherin mediates nuclear-to-cytoplasm transport.
Release of nuclear-imported “cargo” protein exposes nuclear export signal (NES) in importin α.
Importin α-exportin complex binds Ran-GTP (high [Ran-GTP] in nucleus).
Ran-GTP promotes stable assembly of importin α-exportin complex.
Importin α-exportin-Ran-GTP complex transported via NPC into cytoplasm due to Ran-GTP gradient.
![<p><em>How does importin α (transport receptor subunit) move back to the cytoplasm in order to participate in additional rounds of import?</em></p><ul><li><p>Importin α <u>binds</u> to <strong>exportin</strong>.</p><ul><li><p>Karyopherin <u>mediates nuclear-to-cytoplasm transport</u>.</p></li><li><p>Release of nuclear-imported “cargo” protein <u>exposes nuclear export signal (NES)</u> in importin α.</p></li></ul></li><li><p>Importin α-exportin complex binds <strong>Ran-GTP</strong> (high [Ran-GTP] in nucleus).</p></li><li><p><strong>Ran-GTP</strong> promotes <strong>stable assembly</strong> of importin α-exportin complex.</p></li><li><p><strong><u>Importin α-exportin-Ran-GTP</u></strong><u> complex transported via NPC into cytoplasm due to Ran-GTP gradient.</u></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/1c38c333-3367-480d-8fb3-5a667eab5687.png)
nuclear export signal (NES)
Specific stretch/sequence of amino acids recognized by exportin, serve as “zipcode” to mediate targeting of protein from nucleus to cytoplasm.
Several different types; all necessary and sufficient for nucleus-to-cytoplasm targeting.
Most common consists of leucine-rich motif (e.g. -LxxLxxL-).
nucleus-to-cytoplasm transport
Exportin binds to importin α and/or “cargo” proteins exported from nucleus via their NESs.
Release of nuclear-imported “cargo” protein exposes nuclear export signal (NES) in importin α.
Importin α (or NES-containing cargo protein)-exportin complex binds Ran-GTP (high [Ran-GTP] in nucleus).
Ran-GTP promotes stable assembly of importin α (NES-containing cargo protein)-exportin complex.
Importin α-exportin-Ran-GTP complex transported via NPC into cytoplasm due to Ran-GTP gradient.
In cytoplasm, GTP on Ran-GTP hydrolyzed by GAP.
Ran-GDP released from exportin and release of importin α or other NES-containing “cargo” protein.
Importin α: used for another round of import.
Ran-GDP: moves back into nucleus (due to [Ran-GDP] gradient) and converted (via GEF) into Ran-GTP.
Exportin: moves back into nucleus (via exposed NLS and importin) for another round of export.
[Ran-GTP]nucleus > [Ran-GTP]cytoplasm
[Ran-GDP]nucleus < [Ran-GDP]cytoplasm
![<ul><li><p><strong>Exportin</strong> <u>binds</u> to importin α and/or “cargo” proteins exported from nucleus via their NESs.</p><ul><li><p>Release of nuclear-imported “cargo” protein exposes nuclear export signal (NES) in importin α.</p></li></ul></li></ul><ul><li><p>Importin α (or NES-containing cargo protein)-exportin complex binds <strong>Ran-GTP</strong> (high [Ran-GTP] in nucleus).</p></li><li><p><strong>Ran-GTP</strong> promotes <strong>stable assembly</strong> of importin α (NES-containing cargo protein)-exportin complex.</p></li><li><p><strong><u>Importin α-exportin-Ran-GTP</u></strong><u> complex transported via NPC into cytoplasm due to Ran-GTP gradient.</u></p></li><li><p>In cytoplasm, GTP on Ran-GTP hydrolyzed by GAP.</p></li><li><p><strong>Ran-GDP released from exportin and release of importin α or other NES-containing “cargo” protein.</strong></p><ul><li><p><strong>Importin α</strong>: used for another round of import.</p></li><li><p><strong>Ran-GDP</strong>: moves back into nucleus (due to [Ran-GDP] gradient) and converted (via GEF) into Ran-GTP.</p></li><li><p><strong>Exportin</strong>: moves back into nucleus (<u>via exposed NLS and importin</u>) for another round of export.</p></li></ul></li><li><p>[Ran-GTP]<sub>nucleus</sub> > [Ran-GTP]<sub>cytoplasm</sub></p></li><li><p>[Ran-GDP]<sub>nucleus</sub> < [Ran-GDP]<sub>cytoplasm</sub></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/666fc3dc-fa9b-4569-9524-ba37330a8972.png)
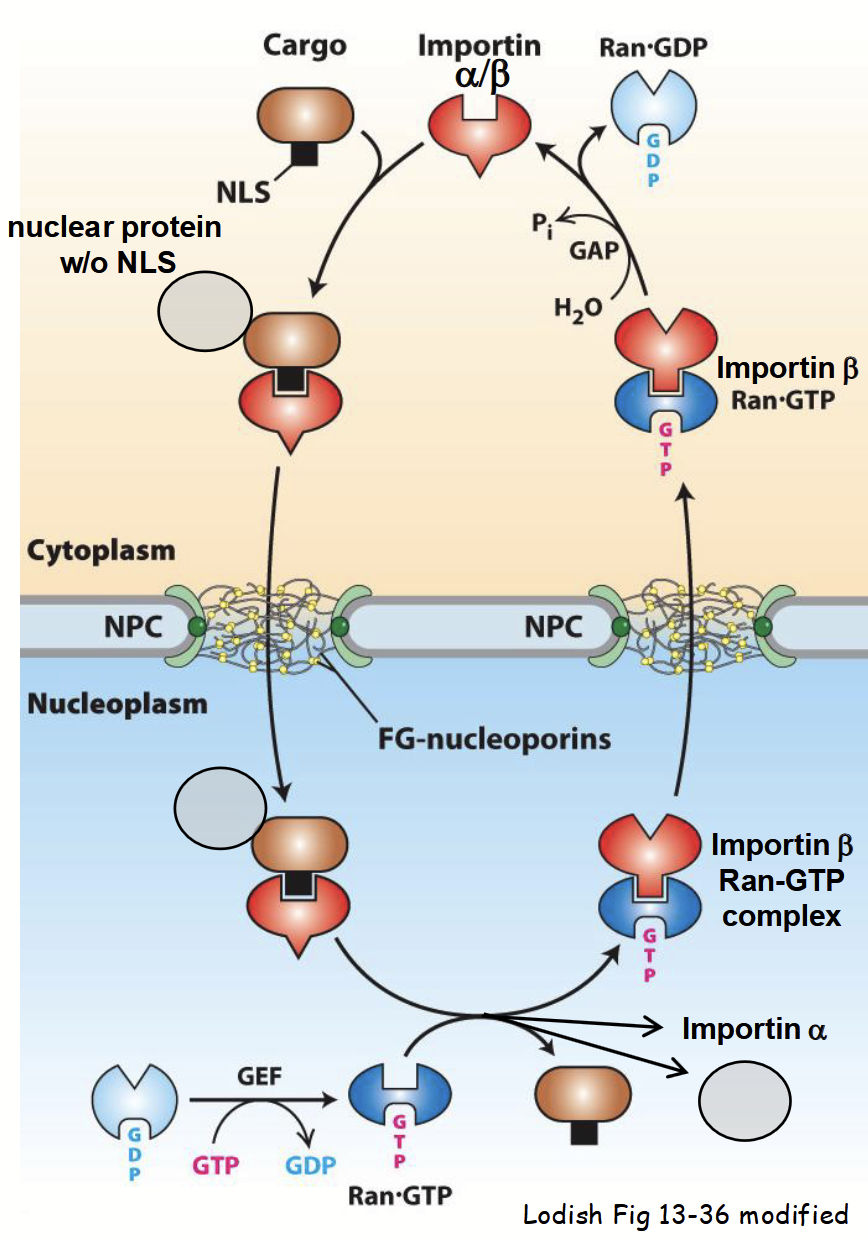
“piggyback” nuclear protein import
Some nuclear proteins have no NLS(s).
Newly-synthesized protein lacking an NLS binds to NLS-containing protein in cytoplasm.
Many proteins “shuttle” between nucleus and cytoplasm - participate in both nuclear and cytoplasmic functions.
Often contain both NLS and NES - relative distribution of protein in either compartment controlled by relative strength of NLS and NES.
e.g. NLS > NES = majority of protein (at steady-state) localized in nucleus.
Can have more than one NLS.
Strength of NLS or NES can be controlled by post-translational modifications.
e.g. phosphorylation of specific AA residue(s) adjacent to targeting signal.
Determines localization: in nucleus or cytoplasm.
Targeting and import of protein-protein complex into nucleus mediated by importin, as usual.
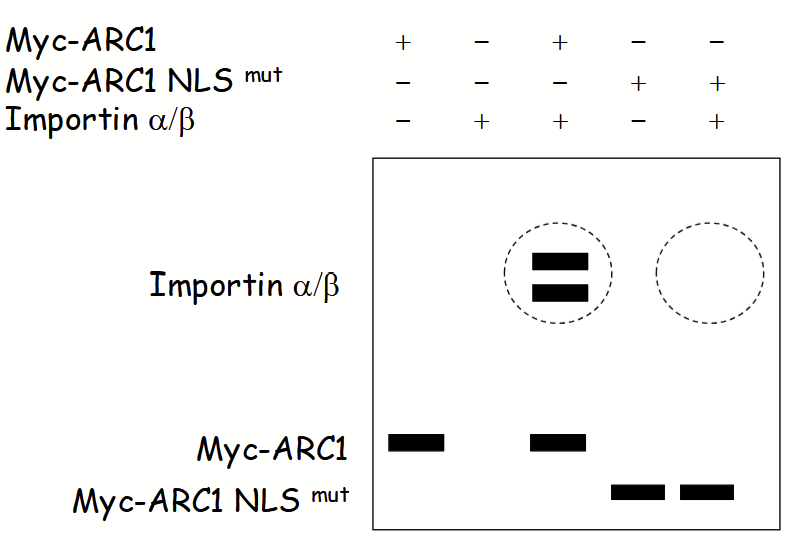
in vitro co-immunoprecipitation assay
Used to assess “cargo” protein-importin binding → does a nuclear-localized protein interact with importin?
“Bait": purified epitope-tagged nuclear protein.
“Prey”: purified importin (alpha/beta subunits).
Step 1: mix “bait” and/or “prey” proteins in vivo.
Step 2: add agarose beads coated with anti-epitope-tag IgGs.
Step 3: isolate beads (via centrifugation) along with all associated (i.e. bait and interacting) proteins.
Step 4: SDS-PAGE and Coomassie blue staining/
cyclins
Nucleocytoplasmic proteins involved in control of cell cycle. Synthesized and degraded during each cell cycle.
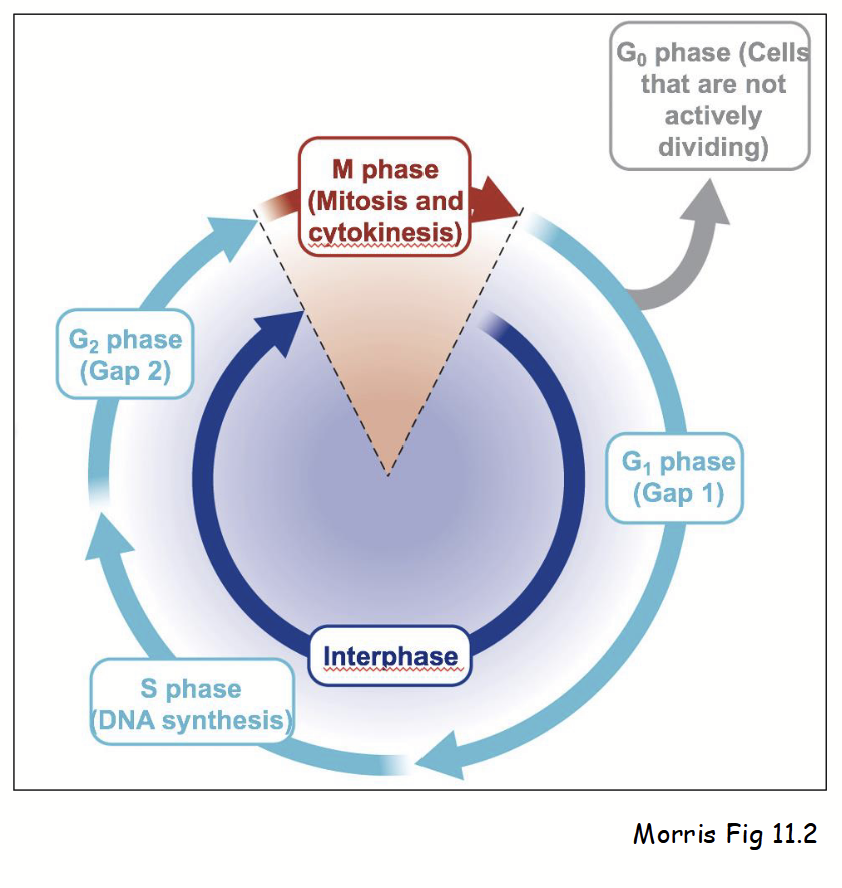
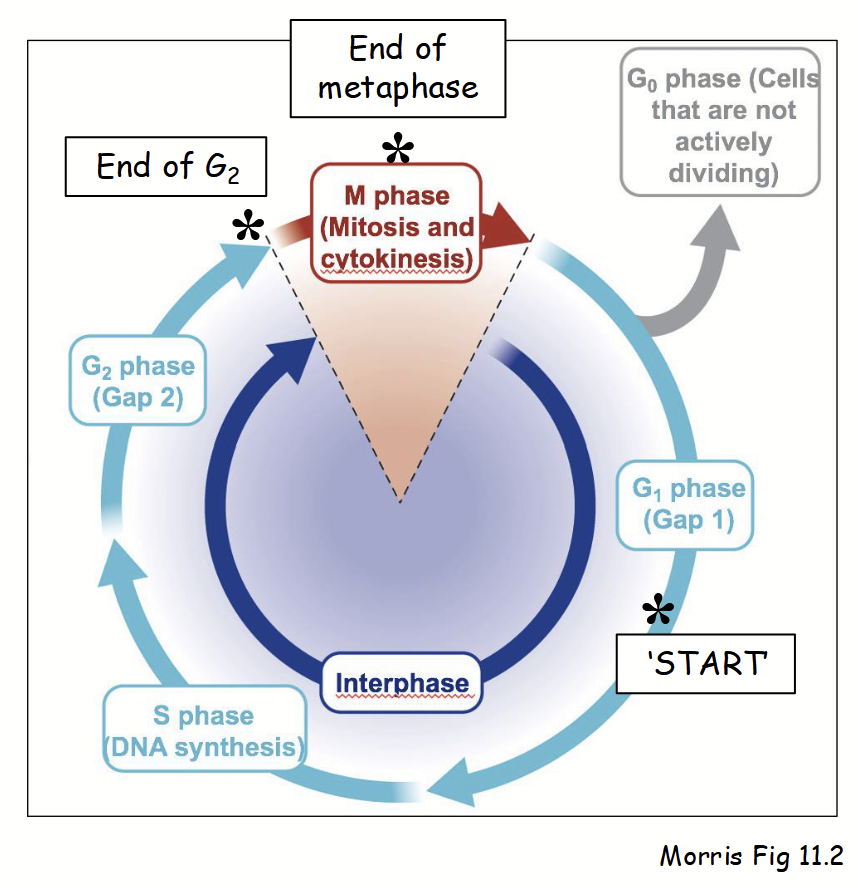
cell cycle
Series of defines stages in cell’s life-cycle. Two main phases:
Interphase (three stages):
G1 (Gap 1): cell performs normal cellular activities and can respond to environment.
S (Synthesis): DNA replication and increased synthesis of factors required for chromosome duplication (e.g. histones).
G2 (Gap 2): cell grows and prepares for mitosis.
Also G0 (Gap 0): non-dividing cells (most cells) - stage when cell “arrests” during G1, rather than proceeding to S.
M phase (mitosis):
Consists of prophase, metaphase, anaphase, and telophase - duplicated chromosomes separated into two nuclei.
Also consists of cytokinesis - mother cell divides into two daughter cells.
Note: prophase involves chromosome condensation, mitotic spindle formation, and reversible breakdown of nuclear envelope (nuclear membranes, lamina, NPCs, etc.).
control of cell cycle
Understanding cell cycle regulation = important implications for cancer.
Progression (or arrest) through cell cycle regulated at distinct stages - “checkpoints”.
Involve surveillance mechanisms that ensure cell cycle proceeds properly.
If not: (e.g. DNA damage due to UV irradiation) cellular signals lead to cell death (apoptosis), cell cycle arrest (senescence), or disease (cancer).
Several primary checkpoints in cell cycle: mid G1, end of G2, end of metaphase.
Progression involves positive controls: nucleocytoplasmic (mobile) factors responsible for mediating transitions from one phase to next.
Transition through checkpoints controlled by mitotic cyclins and cyclin-dependent kinases (CDKs).
cancer
Inability of cell to regulate division (i.e. uncontrolled cell proliferation).
mid G1 checkpoing
“Restriction point” or “START” point in the cell cycle: cell commits to DNA replication in S and organelle duplication begins.
end of G2 checkpoint
Cell cycle checkpoint in which the cell commits to entering M phase.
end of metaphase
Cell cycle checkpoint in which the cell commits to chromosome segregation.
cyclin-dependent kinases (CDKs)
Cell-cycle-specific kinase enzymes located in the nucleus; phosphorylate various “target” (nuclear) proteins - turn “on” or “off”.
cyclins
Nucleocytoplasmic proteins; bind CDKs and regulate their activity during specific stages of cell cycle.
e.g. mitotic forms during G2-to-M transition.
cyclins concentration
The abundance of cyclin proteins - varies in a cyclical fashion during cell cycle.
[Mitotic cyclins] varies during the cell cycle:
Early interphase (G1): [mitotic cyclins]low → CDK activitylow
End of G2/start of M phase: [mitotic cyclins]high → CDK activityhigh
Results in phosphorylation of “target” proteins and cell enters M phase.
CDK “target” proteins in nucleus at end of G2 and start of M phase
Histones and condensins: phosphorylation leads to chromatin packing and chromosome condensation in preparation for mitosis.
Lamins: phosphorylation leads to disassembly of nuclear lamina.
Nups (nucleoporins): phosphorylation leads to disassmbly of NPCs.
histones and condensins
Target proteins in nucleus at the end of G2 and start of M phase. Phosphorylation leads to chromatin packing and chromosome condensation in preparation for mitosis.
lamins
Target proteins in nucleus at the end of G2 and start of M phase. Phosphorylation leads to disassembly of nuclear lamina (lose their ability to interact with each other).
nucleoporins at end of G2 and start of M phase
Target proteins in nucleus at the end of G2 and start of M phase. Phosphorylation leads to disassembly of NPCs.
“open mitosis”
Nuclear structure during mitosis.
Dynamic process in higher eukaryotes: nucleus completely disassembled by metaphase.
Prophase: outer and inner nuclear membranes break down, lamina and NPCs disassemble, membrane-bound and soluble nuclear proteins “released” into ER membrane and cytoplasm, respectively.
Two daughter nuclei reassemble during end of mitosis (i.e. telophase - after chromosomal division).
[Mitotic cyclin]low → CDK activitylow → dephosphorylation of Nups and lamins → reformation of nuclear lamina, nuclear envelope and NPCs → re-import of soluble NLS-proteins from cytoplasm.
oscillations in [cyclin]
Control of the cell cycle
Occur due to relative rates of protein synthesis and degradation at different points during the cell cycle.
Decrease in [mitotic cyclin] after start of M phase due to decreased synthesis of new cyclin proteins and degradation of pre-existing cyclin proteins (via proteasome).
CDKs inactivated by phosphorylation by CDK kinases, CDK kinase kinases, etc. (complex regulatory process).
AND… after start of M phase, pre-existing cyclins also prevented from targeting to nucleus… (cannot activate CDKs in nucleus).
![<p><strong>Control of the cell cycle</strong></p><p>Occur due to relative rates of protein synthesis and degradation at different points during the cell cycle.</p><ul><li><p>Decrease in [mitotic cyclin] after start of M phase due to decreased synthesis of new cyclin proteins <u>and</u> degradation of pre-existing cyclin proteins (via proteasome).</p></li><li><p>CDKs inactivated by phosphorylation by CDK kinases, CDK kinase kinases, etc. (complex regulatory process).</p></li><li><p>AND… after start of M phase, <u>pre-existing cyclins also prevented from targeting to nucleus…</u> (cannot activate CDKs in nucleus).</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/71447631-7d92-4273-aad8-dd3e02aba686.png)
nucleocytoplasmic transport of cyclins
Cyclins normally shuttle between nucleus and cytoplasm - possess both NLS and NES.
Relative strength of targeting signal(s) results in cyclin protein localized primarily in cytoplasm and/or nucleus.
e.g. localization of the mitotic cyclin B1 in mammals:
Up to and during G2: cycling shuttles between nucleus and cytoplasm, but localizes primarily in cytoplasm (NES > NLS).
End of G2/start of M phase: NES in cyclin phosphorylated; NLS > NESPhos … localizes primarily in nucleus; activate CDKs.
After start of M phase: NES in cyclin dephosphorylated; NES > NLS … localizes primarily in cytoplasm.
standard brightfield microscope
Main components: light source, condenser lens, stage (holding specimen), objective and ocular (projection) lenses, and “detector” (eye).
Light diffracted by specimen; undiffracted light (e.g. field-of-view) focused by objective lens.
Image usually captured by video camera:
More sensitive to low light intensities - living cells can be viewed with limited photo (light) damage.
Record image as digital file - different light intensities converted into 2D array of numbers (quantified).
Easily manipulate digital images using various computer software programs:
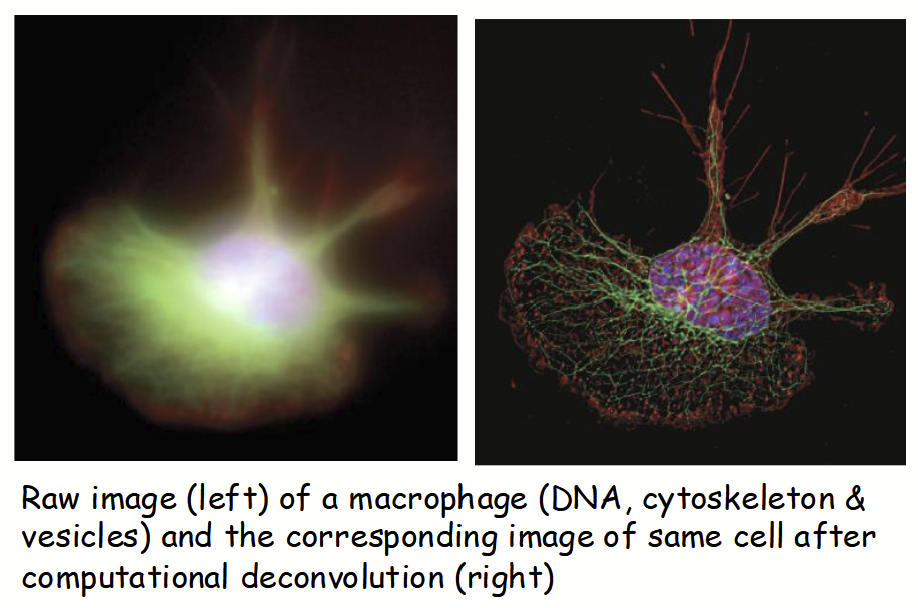
e.g. deconvolution - designed to remove background and out-of-focus light (yields high contrast and clarity).
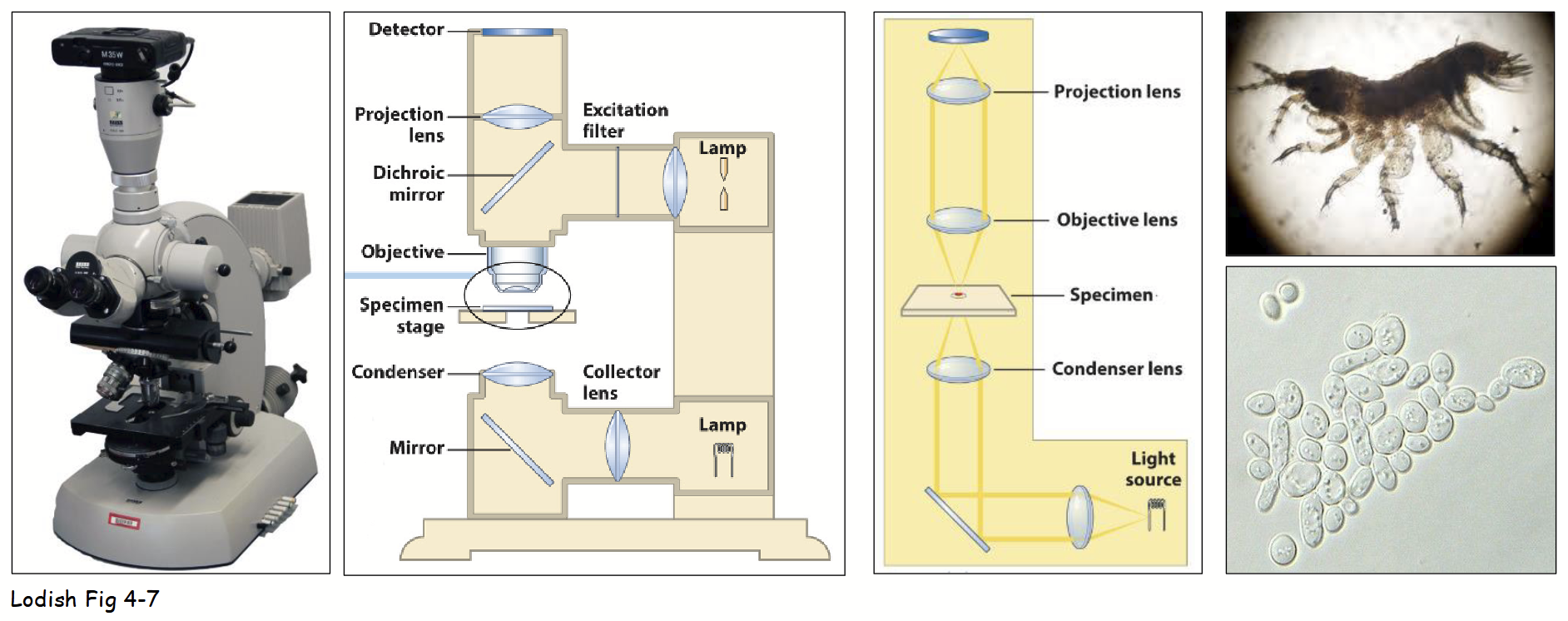
deconvolution
Standard brightfield microscope - designed to remove background and out-of-focus light (yields high contrast and clarity).
magnification
Primary purpose of microscopy: generate magnified, high-quality view of speciman.
Overall magnification: objective lens x ocular lens.
Quality of image - does continuing to enlarge image provide more details/information?
No: “empty” magnification.
overall magnification
= objective lens x ocular lens
empty magnification
Continuing to enlarge the image does not provide more details/information about the speciman.
resolution
The most important aspect of today’s microscope.
Minimum distance that can separate two points that still remain identifiable as separate points.
i.e., ability to distinguish two close objects as separate entities.
resolving power
Depends on two main factors in microscopy:
Wavelength (λ) of illumination light.
Numerical aperture (NA): light-gathering qualities of objective lens and specimen mounting medium.
resolution
The distance in nm = (0.61 x λ) / NA
Maximized by:
Use of shorter λ of illuminating light.
e.g. blue light < red light
Increase NA - alter mounting medium (e.g. air → oil).
Limit for most standard brightfield (and CLSM) is ~200 nm.
Can only observe larger organelles (e.g. nuclei, mitochondria, chloroplasts)
numerical aperture (NA)
Light-gathering qualities of objective lens and specimen mounting medium.
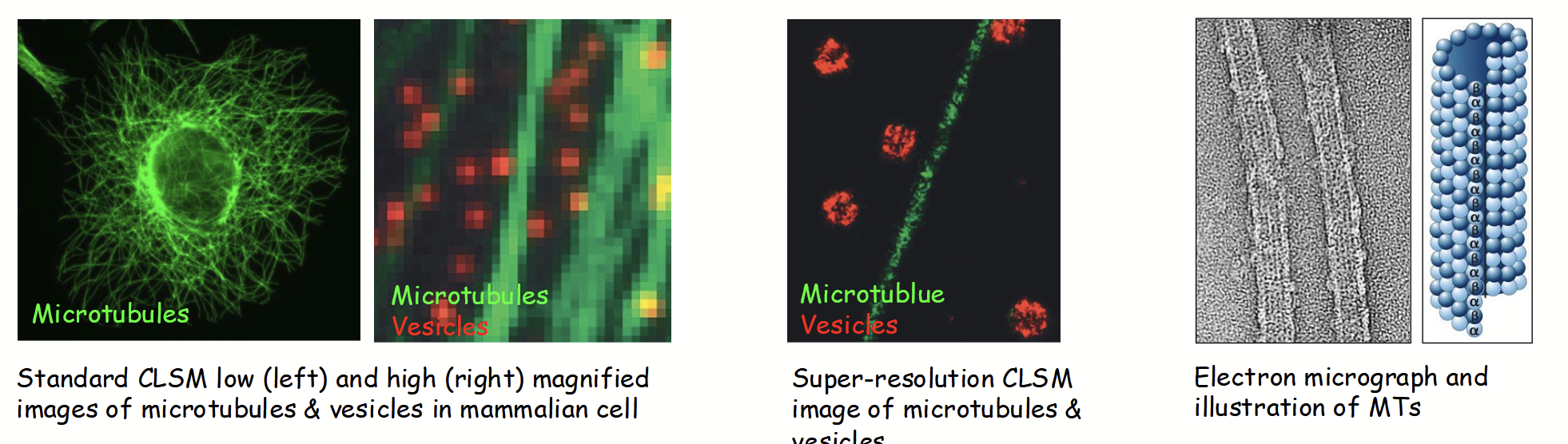
limits of resolution
Human eye: 0.1 mm
Standard brightfield and fluorescence microscopy (CLSM): 200 nm (500X)
Super-resolution CLSM: 20 nm (5,000X)
Electron microscope (EM): 0.2 nm (500,000X)
Uses electrons (λ=0.0045 nm) rather than photons - lower λ yields higher resolution.
brightfield microscopy limitations
Major: speciman’s poor contrast (i.e. lack of structural/cellular details).
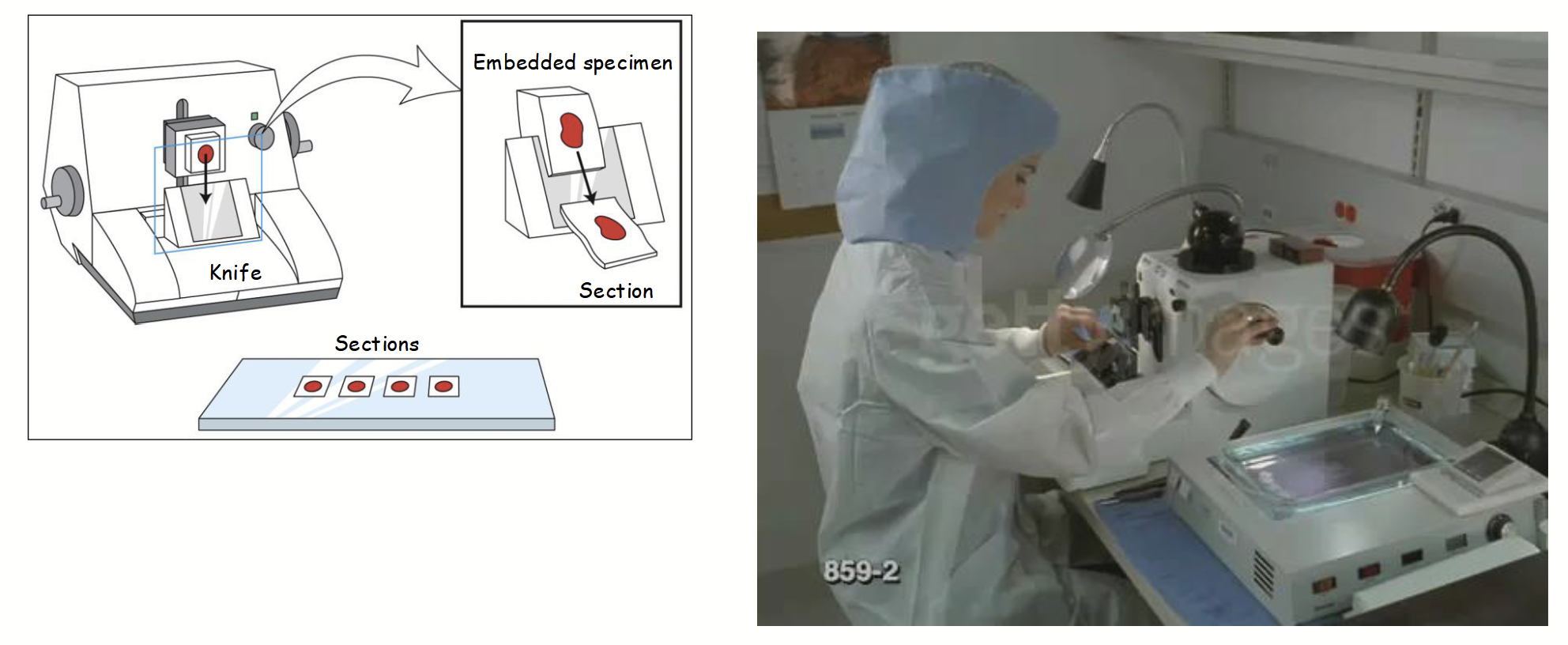
Specimens usually “fixed”, embedded (in plastic/wax) for support, then sectioned with microtome, and stained with molecule-specific dye(s).
e.g. formaldehyde fixation: cross-links amino groups on adjacent proteins/nucleic acids.
CON: fixation results in cell death, embedding and sectioning and can lead to structural “artifacts”, and limited (molecule-specific) stains.

speciman sample sectioning using a microtome
fluorescence microscopy
Microscopy technique for visualizing fluorescent molecules in living (or fixed) specimens.
Relies on endogenous fluorescence in specimen (autofluorescence), applied fluorescent dyes or dye-conjugated antibodies (immunogfluorescence), and/or autofluorescent proteins.
PROS: provides increased contrast and allows study of structure(s) and (when not fixed) dynamic processes in living cells, and in 3D.
CONS: out-of-focus fluorescence from (thick) specimen results in “blurred” image.
Various methods, including Confocal Laser-Scanning Microscopy (CLSM).
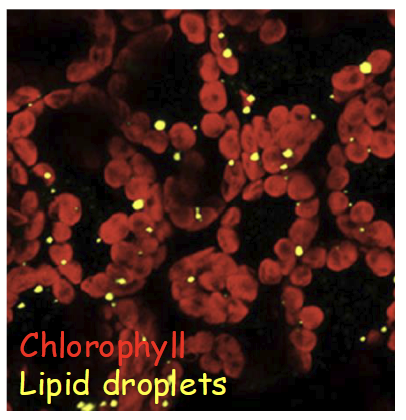
autofluorescence
Type of fluorescence microscopy that relies on endogenous fluorescence in the specimen.
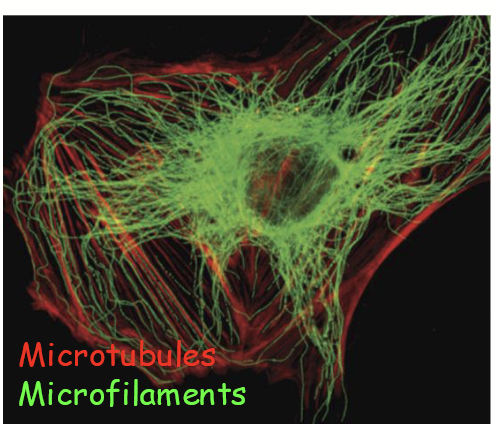
immunofluorescence
Type of fluorescence microscopy that relies on dye-conjugated antibodies.
autofluorescent proteins

fluorescence microscopy pros
Provides increased contrast and allows study of structure(s) and (when not fixed) dynamic processes in living cells, and in 3D.
fluorescence microscopy cons
Out-of-focus fluorescence from (thick) specimen results in “blurred” image.
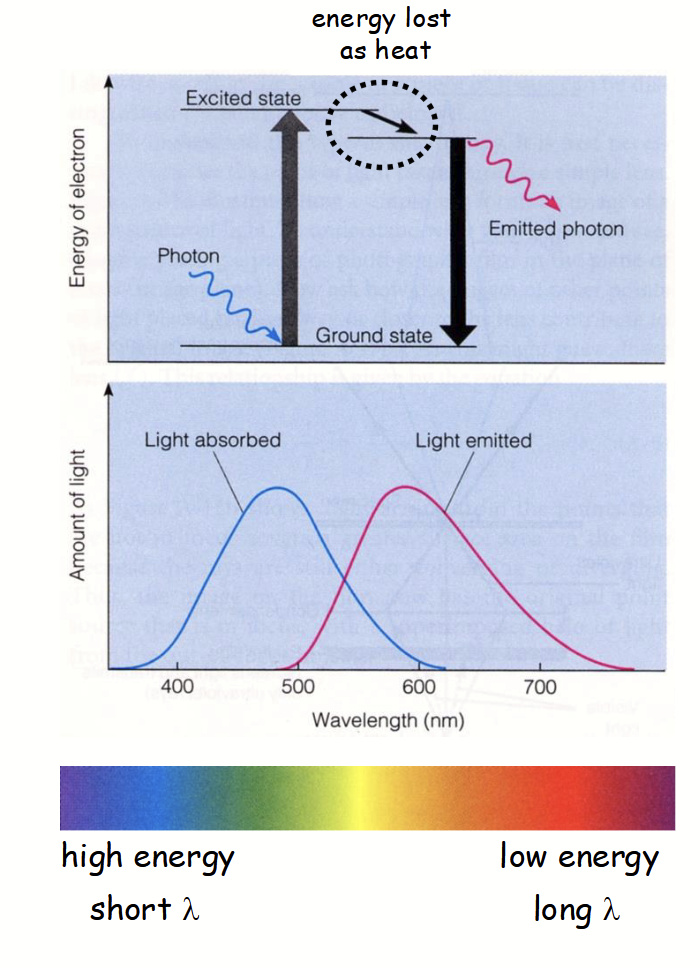
principles of fluorescence
Complex process in fluorescence microscopy
Certain atoms in fluorescent molecule (e.g. GFP) can absorb photon of certain λ (e.g. blue light).
Atom’s electron becomes “excited” and moves up to higher energy state.
“Excited” electron is highly unstable.
It loses energy and returns to “ground state” by emitting a photon with lower energy (i.e. longer λ; e.g. red light).
“Emitting” electron has lower energy (longer λ) because some energy is initially lost as heat.
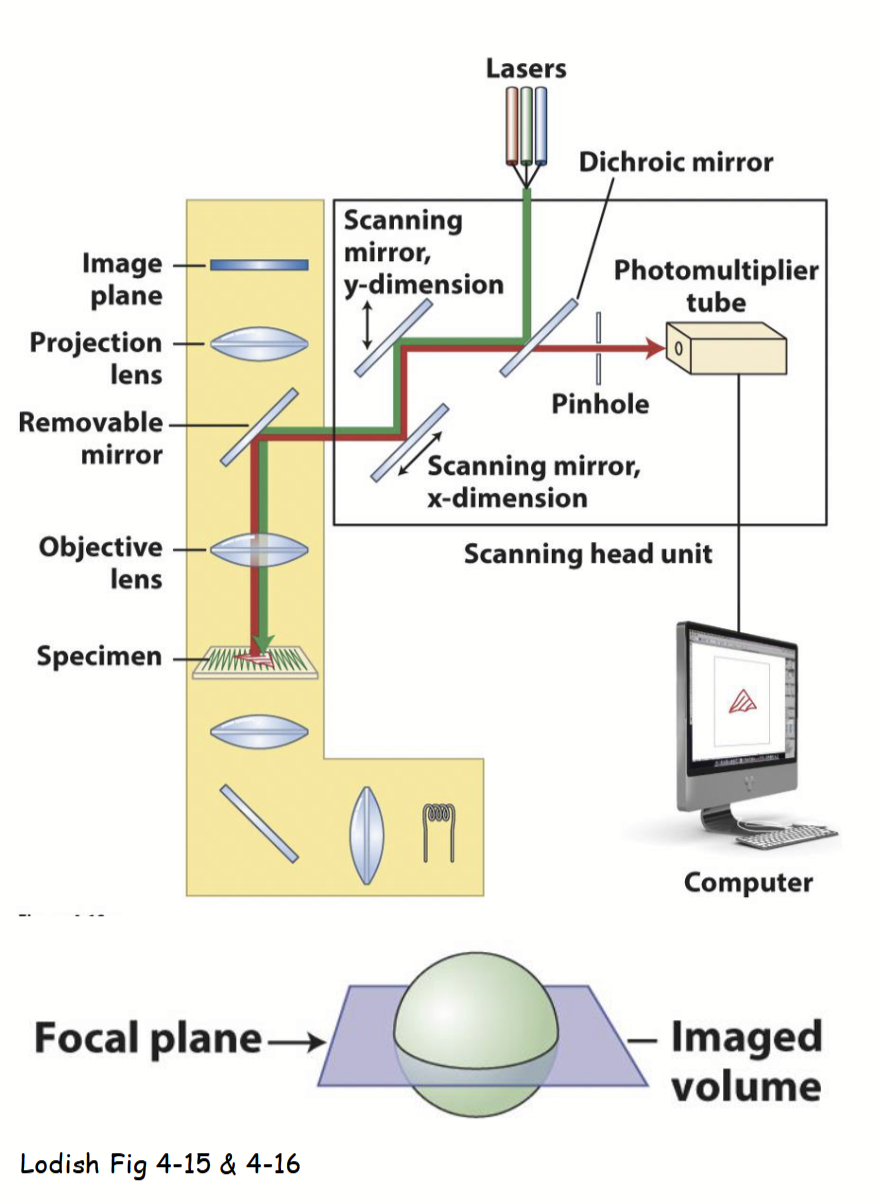
Confocal Laser-Scanning Microscopy (CLSM)
Method of fluoresence microscopy that has a similar set-up to a standard brightfield light microscopy, but with additional features.
One or more lasers of certain wavelengths of light “excite” fluorescent molecules in the specimen, emitted light is specifically focused to obtain a detailed image.
Speciman viewed using this method is usually living (not necessary to “fix” speciman).
Allows for viewing dynamic biological/cellular processes live - via endogenous autofluorescence, vital fluorescent dye, or ectopically-expressed autoflorescent fusion protein.
Lasers can penetrate further into thicker living specimens (compared to standard light microscopy).
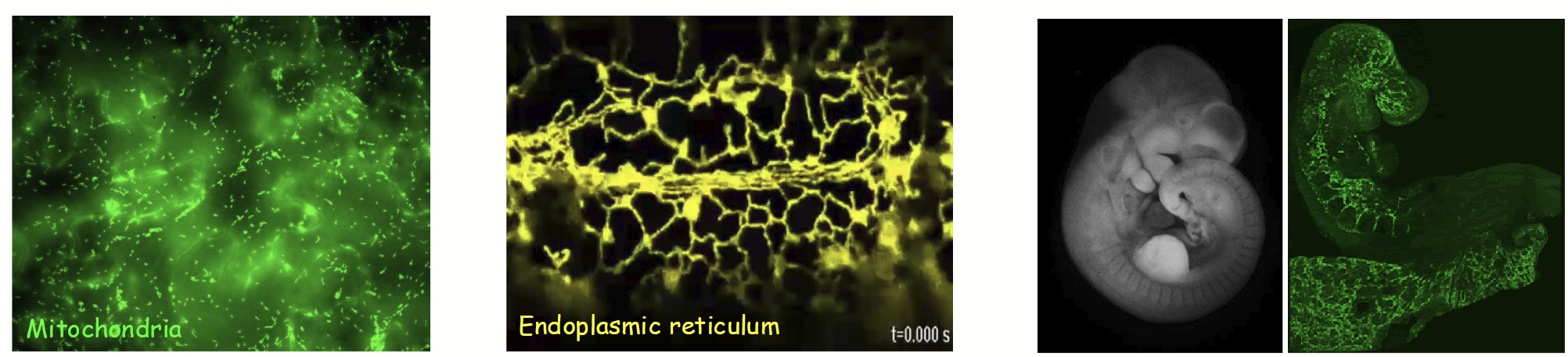
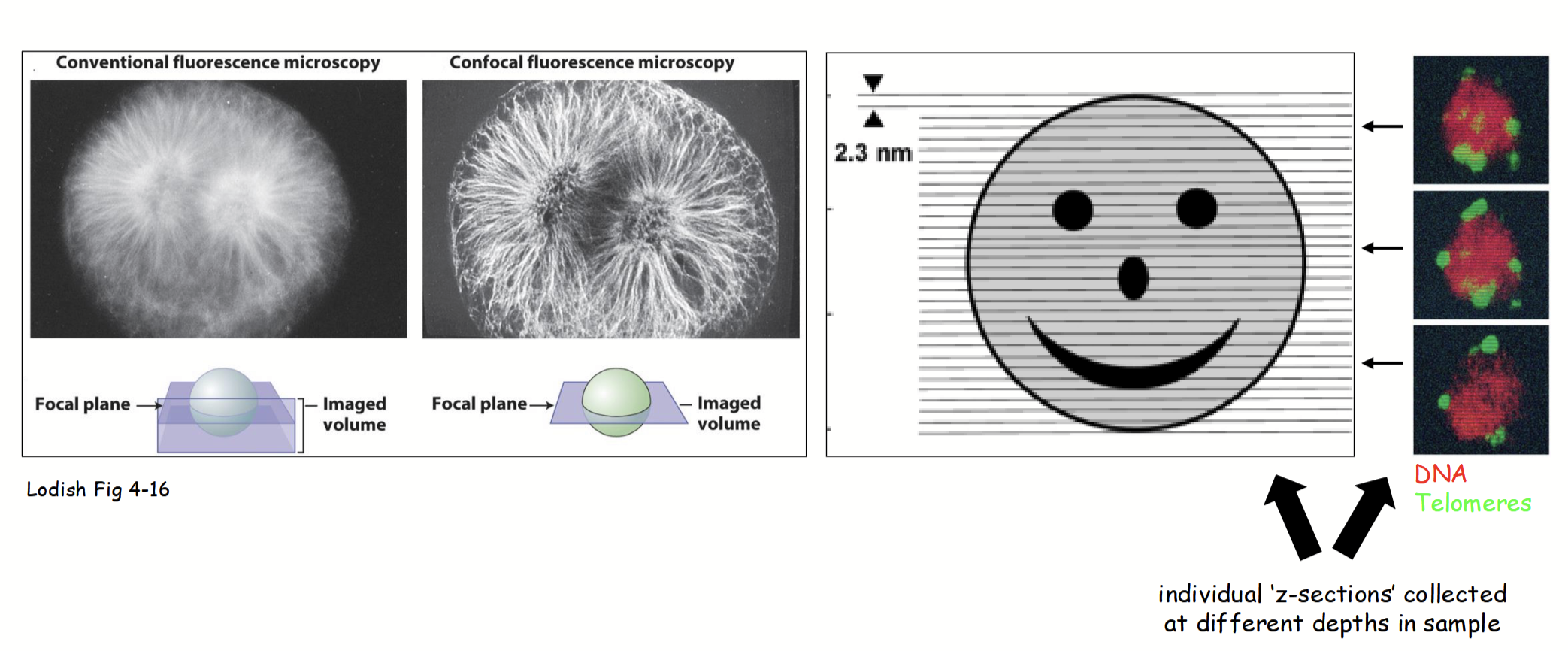
CLSM process
Speciman rapidly scanned with point laser light at a specific excitation λ (based on fluorescence of molecule(s) being detected).
Emitted fluorescent light from only a single layer (focal plane) within the specimen is focused through the pinhole and then collected/viewed.
All out-of-focus fluorescence from the specimen (i.e., emitted light from above and below the focal plane) is excluded (does not pass through pinhole).
Yields an individual 2D z-section (“optical slice”) of the specimen that is less “blurry” than images obtained with standard fluorescence microscopy.
Individual “z-sections” collected at different depths in sample and combined (i.e., z-sections stacked together serially) to form z-stack and generate 3D image.
z-section
In CLSM, the yielded individual 2D “optical slice” of the specimen that is less blurry than images obtained with standard fluorescence microscopy.
Individual parts are collected at different depths in sample and combined to form a stack and generate a 3D image.