Final Exam learning objectives
Know the difference between prokaryotic and eukaryotic cells and where processes of the central dogma take place
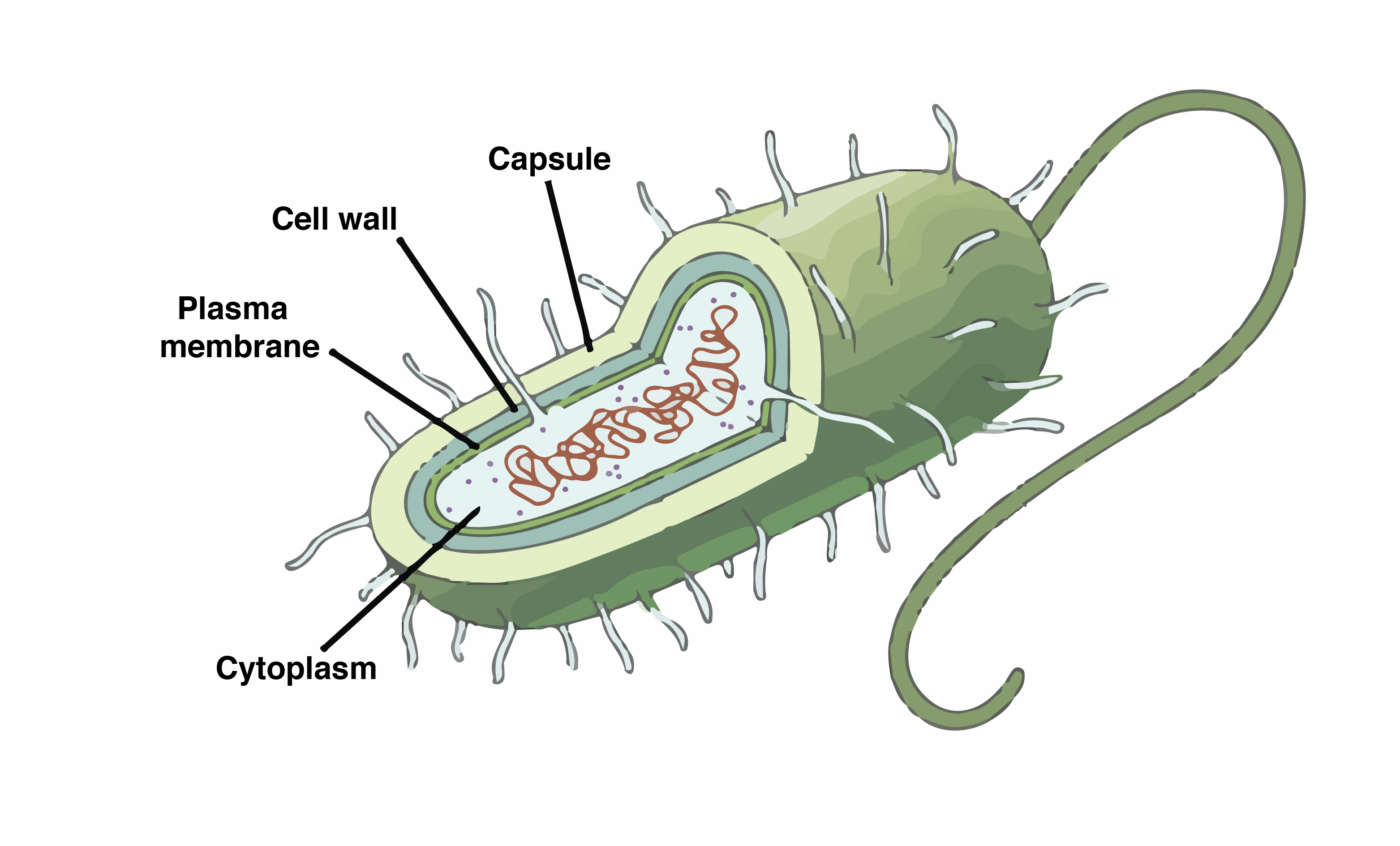
Prokaryotic Cells-
All processes happen in the cytoplasm
Eukaryotic Cells-
DNA replication- Nucleus
Transcription- Nucleus
translation- Cytoplasm

Know the properties of water
Water is polar (partial charges on H and O)
Hydrogen bonding (allows water to form strong intermolecular interactions
Known as a universal solvent
Cohesion and adhesion- important for biological transport
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
Know the difference between prokaryotic and eukaryotic cells and where processes of the central dogma take place
Prokaryotic Cells-
All processes happen in the cytoplasm
Eukaryotic Cells-
DNA replication- Nucleus
Transcription- Nucleus
translation- Cytoplasm


Know the properties of water
Water is polar (partial charges on H and O)
Hydrogen bonding (allows water to form strong intermolecular interactions
Known as a universal solvent
Cohesion and adhesion- important for biological transport
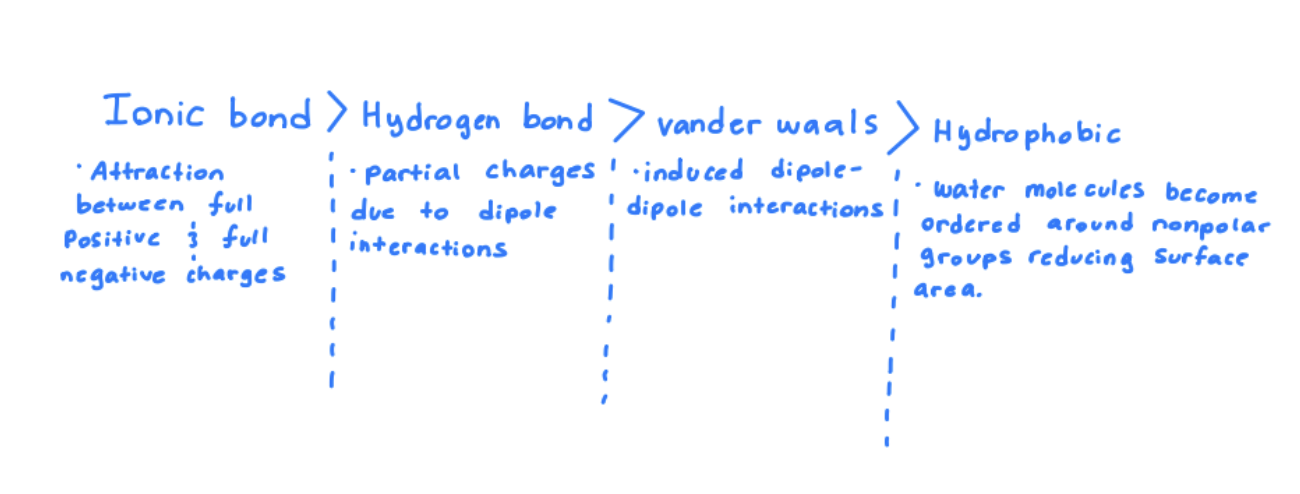
Describe the weak interactions that are central to biology and rank from strongest to weakest
Define base and acid
Acid: A substance that donated a proton (H+) in solution
Ex: HCl —→ H+ + Cl-
Base: A substance that accepts a proton (H+) or donates OH- in solution
Ex: NH3 + H+ —→ NH4+
conjugate base: Whats left after an acid donates a proton. For example an acid can be NH4+ and its conjugate base can be NH3
Know and be able to use the Henderson-Hasselbach equation
The Henderson-Hasselbach equation explains the relationship between PH and pKa
PH= pKa + log (conjugate base / acid)
Know your amino acids: structure (recognize), single-letter code, three-letter code.
Play amino acid quiz game
Draw Fischer and stereochemical representations of amino acids
Know types of functional groups of amino acids and their properties
(mnemonic) For hydrophobic
GAVLIMP
Glycine
Alanine
Valine
Leucine
Isoleucine
Methionine
Proline
(mnemonic) For aromatic
WYF
Tryptophan
Tyrosine
Phenylalanine
(mnemonic) For Hydrophilic
STQNC
Serine
Threonine
Glutamine
Asparagine
Cysteine
(mnemonic) For Negative charge/acidic
Ed
Glutamic acid
Aspartic acid
(mnemonic) For Positive charge/basic
HKR
Histidine
Lysine
Arginine
Know properties of amino acids in physiological pH and how pH changed affect these properties, know pKa
Explain what an essential amino acid is
An essential amino acid is one that cannot be made by the body and must be obtained from the diet
Required for protein synthesis
Compare different levels of protein structure and how they relate to each other
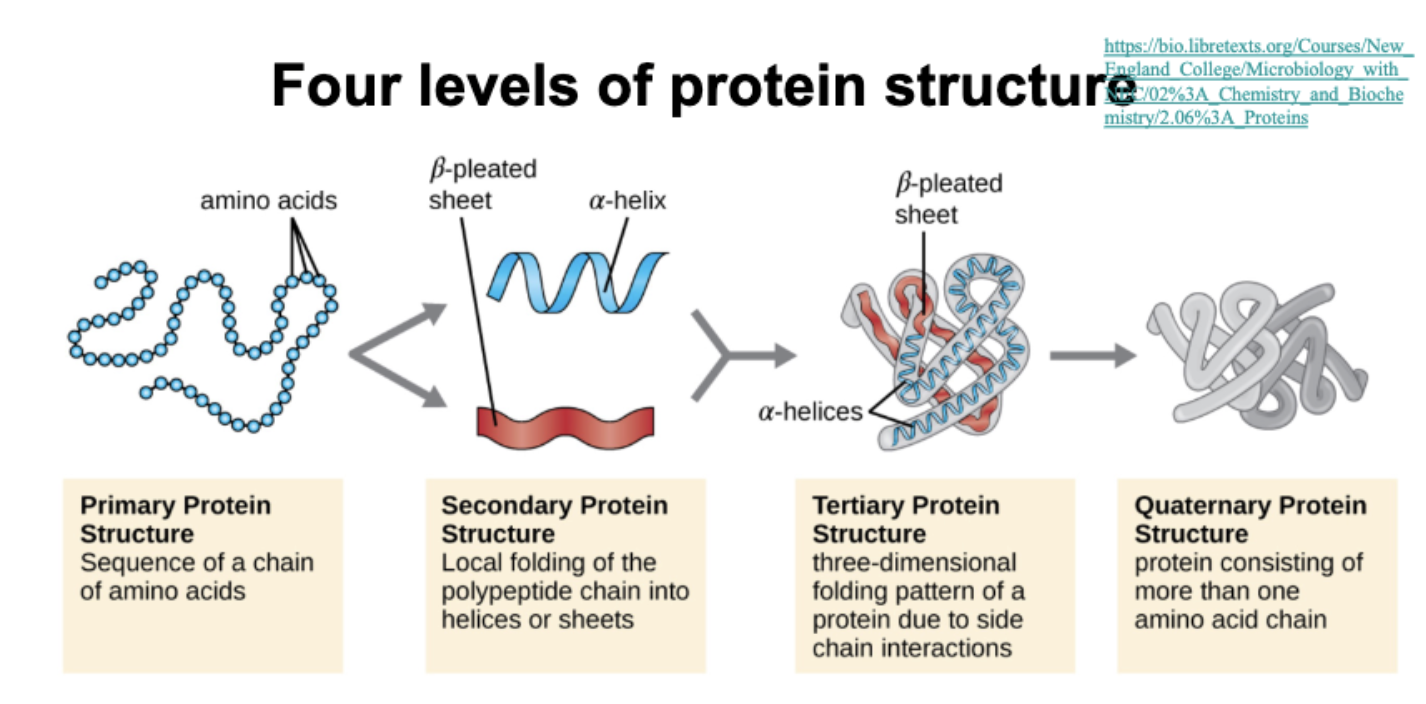
Basic understanding of how proteins fold
Folding is guided by the amino acid sequence and stabilized by non-covalent interactions
Primary structure: amino acid sequence
Secondary structure: Local folding into alpha-helices and beta-sheets via hydrogen bondings
Tertiary structure: 3D folding driven by hydrophobic interactions, hydrogen bonds, ionic bonds, and disulfide bridging
Quaternary structure: Multiple polypeptides assemble into a function protein
Describe the fundamental properties of peptide bonds
Bond between a Carboxylic acid group of one amino acid and NH2 of another amino acid forming a peptide bond that has partial double bond characteristics because of resonance.
planar and rigid so it doesn’t rotate freely
Trans configuration favored
Describe the significance of the Ramachandran plot
organizes possible steric combinations and tells us about secondary structure of an amino acid
Shows allowed angles of rotation for the backbone of a protein
(phi= rotation around N-alpha carbon bond)
(psi=rotation around alpha carbon-Carbon bond
Describe what drives protein folding
Entropy driven by hydrophobic effect, non-polar side chains avoid water becoming buried inside the folded protein structure.
Explain the conclusions of Anfinsen’s experiment
Most important outcome of Anfinsen’s experiment was that the primary amino acid sequence of a protein determines its three-dimensional structure
Anfinsen ran an experiment using the denaturing agents Urea and B-mercaptoethanol that focused on an enzyme called ribonuclease. In which he planned to destroy the tertiary structure and investigate which proper conditions formed the tertiary structure
Experiment #1- Excess BME and 8M urea were added together where the outcome was a denatured enzyme. Once the reagents were both removed the enzyme eventually reformed the original tertiary structure.
Experiment #2- Excess BME and Urea were added together where first B-merca was removed and then urea where then a scrambled enzyme was seen with improper di-sulfide bonds
Experiment #3- Trace amounts of BME were added to the scrambled enzyme where the correct tertiary structure was re-formed
The experiments also showcased how the protein folding is a reversible process and is sequence driven where the right folding needs to happen before stabilization bonds.
Describe chromatography techniques for protein purification
Affinity Chromatography
Separates based on specific binding interactions like His-tagging (highly selective and efficient)
Ion-Exchange Chromatography
Separates by charge
Proteins bind to a charged resin (positive or negative) and are eluted by changing salt concentration or pH
Size-Exclusion Chromatography
Separates proteins by size
Large proteins elute first because they don’t enter the pores of the beads
Describe the principles of separation by SDS-PAGE
SDS-PAGE separates proteins based on their molecular weight
SDS (sodium dodecyl sulfate) coats proteins with negative charge and denatures them (removes shape differences)
Proteins lose their shape and charge differences, separation is then based on only size
Proteins migrate through polyacrylamide gel, where smaller proteins move faster
Resulting in a band on a gel that reflects protein size, not shape or charge
SDS-page separates proteins by size only, because SDS gives them a uniform charge and shape
Describe the use of an assay
An assay is used to measure the presence, activity, or concentration of a specific molecule such as an enzyme or protein in a sample
Define what an enzyme is
A biological catalyst, usually a protein; that speeds up chemical reactions in living organisms without being consumed in the process
It works by lowering the activation energy, allowing reactions to occur more efficiently and at faster rates
Define how enzymes function
substrate binds to the enzymes active site
The enzyme converts substrate to product, stabilizing the transition state
Product is released from the enzyme allowing it to be re-used for more reactions.
Define the role of free energy in catalysis
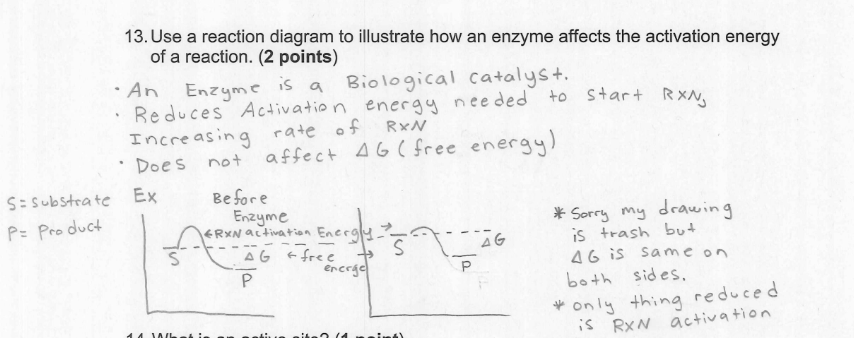
Free energy in catalysis refers to lowering the activation energy allowing the reaction to proceed quicker, without changing the overall free energy of the reaction
Define standard free energy
Standard free energy measures how much energy is released or needed by a reaction under set, standard conditions (1M conc. for all reactants and products, 25 celsius (298K), and 1 atm pressure
Define the role between a transition state and an enzyme
Enzymes stabilize the substrates transition state lowering the activation energy and speeding up the reaction
List the features of an active site
the active site is the region on the enzyme where catalysis occurs
Some of the features of an enzyme’s active site include
Specificity- Binds only to a specific substrate due to shape and chemical complementarity
Binding site- Holds the substrate in place through non-covalent interactions like hydrogen bonds, ionic bonds, and van der waals forces
Catalytic residues- Amino acids within the site that participate in the chemical reaction
Induced fit- Can change shape slightly to better fit the substrate upon binding
Transition state stabilization- that helps lower the activation energy by stabilizing the high-energy intermediate
Describe the two models of substrate binding
lock and key model: Active site has a specific shape that exactly fits the substrate. Like a key fitting into a lock. Emphasizing specificity.
Induced fit model: The active site changed slightly to better fit the substrate. More flexible than a lock and key, helping stabilize the transition state for a reaction.
Know what substrate state inhibitors mimic
mimics the transition state of a substrate
Enzymes bind the transition state most tightly blocking the real substrate, tricking the enzyme into binding them
List the assumptions made for Michaelis-Menten kinetics
Steady state assumption; the concentration of ES remains constant over time
Enzyme-substrate complex (ES) forms rapidly and reversibly over time
Initial velocity is measured, product formation is measured early in the reaction, so product does not go back to substrate (irreversible)
Define KM and VMAX
V-max= Max point where the enzyme can’t help increase rate of reaction anymore becoming saturated (all enzyme active sites are occupied)
KM= The substrate concentration at half of V-max where it shows enzymes affinity for substrate
Vo= Enzyme just meets substrate and no product has been built yet measures how fast product is made right in the beginning.
Define catalytic efficiency
Measure of how efficiently an enzyme converts a substrate into product
Draw the [S] vs V0 curve for Michaelis-Menten enzymes and describe the significance of its shape
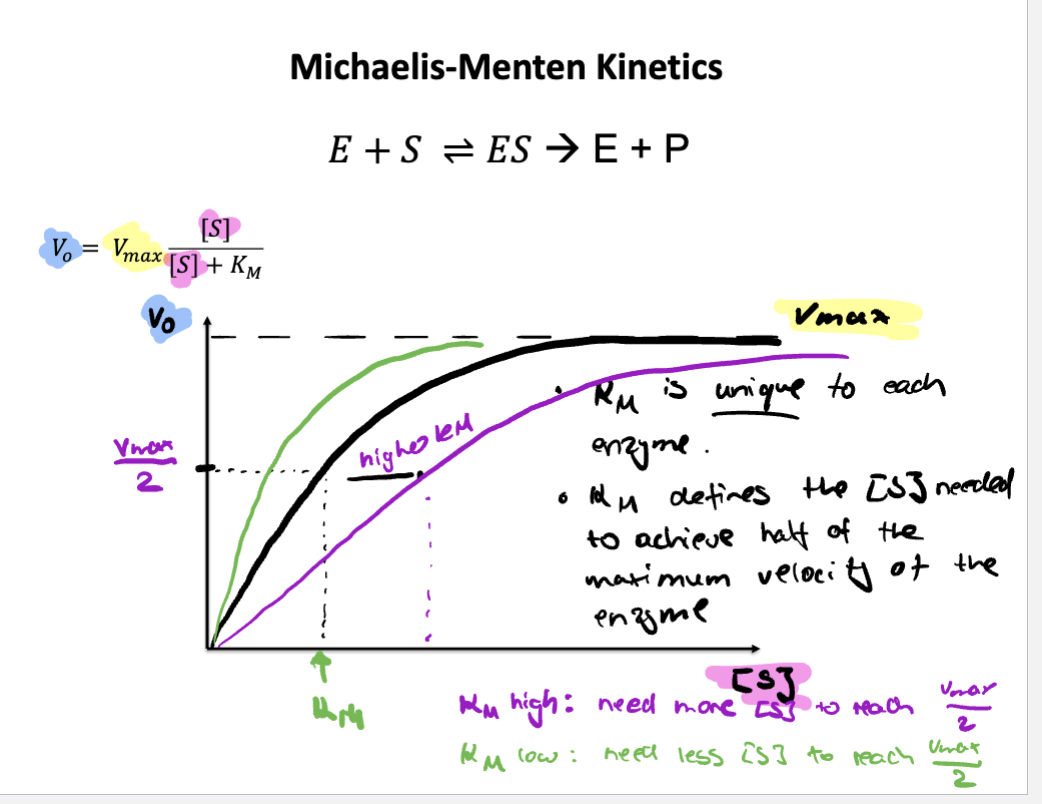
curve is hyperbolic
Typical curve of non-cooperative enzymes
Hyperbolic shape shows saturation behavior. At low [S] the curve rises steeply, the enzyme has plenty of free active sites, so adding more substrate quickly increases the reaction rate. At high [S] the curve levels off because the enzyme becomes saturated meaning all the active sites are occupied, adding more substrate won’t increase the rate much.
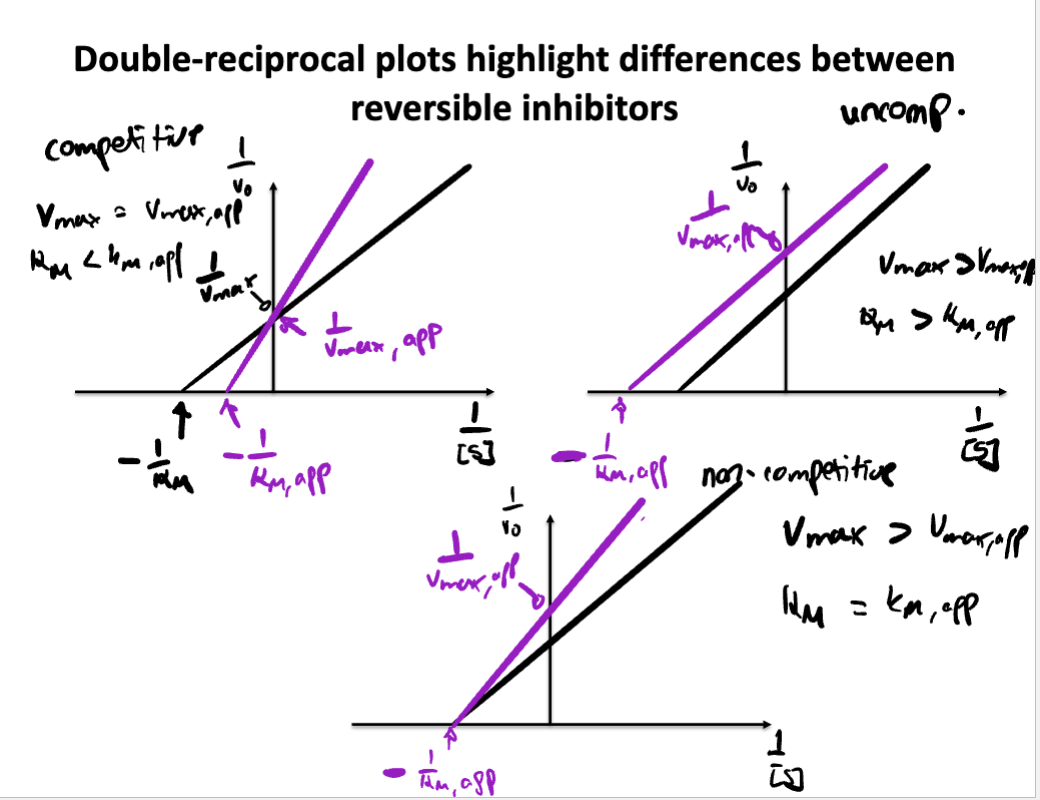
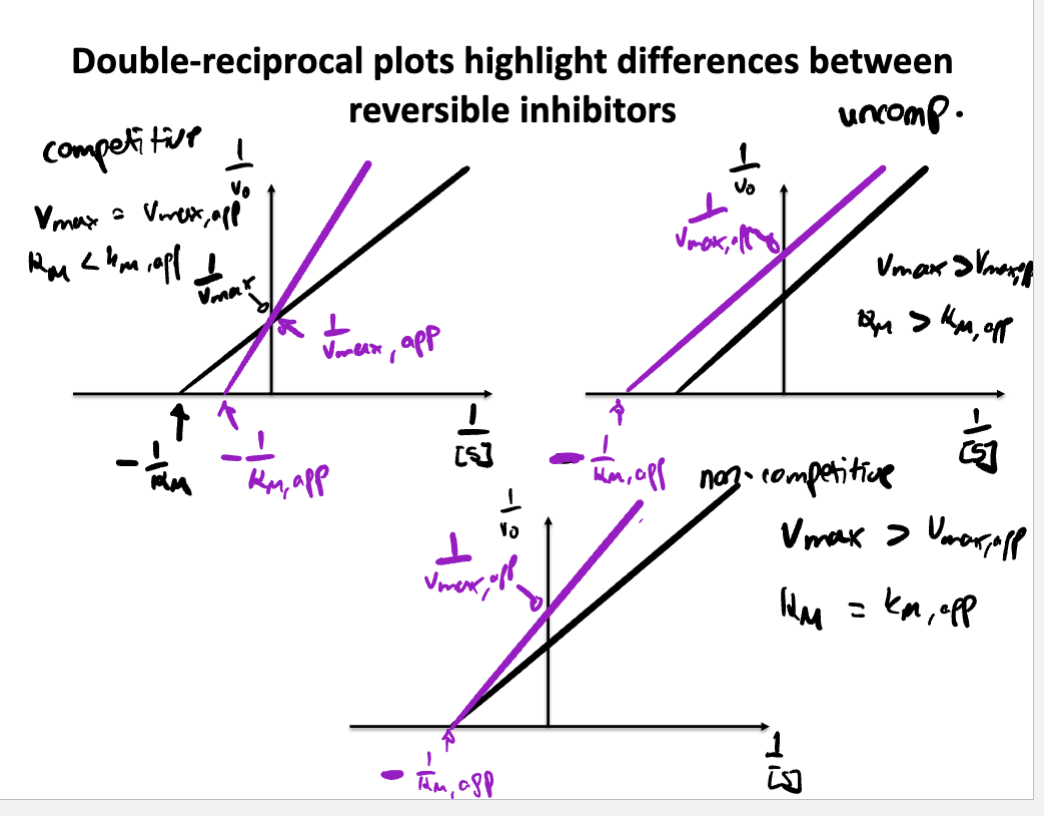
Describe/Draw a lineweaver-Burk plot and explain how different inhibitors change the plot compared to an enzyme without inhibitor
Define different catalytic strategies
Acid-base catalysis: enzyme donates or accepts protons to stabilize transition state.
Covalent catalysis: enzyme forms a temporary covalent bond with the substrate
Metal Ion catalysis: Metal ions stabilize charge or assist in redox reaction
Catalysis by approximation: Brings two or more substrate into close proximity and correct orientation within active site to increase chance of reaction
Define the effect of temperature and pH on enzyme function and make predictions based on active site composition
Effect of temperature:
Increases reaction rate up to an optimal temperature
too high= enzyme denatures (loses/shape function)
Effect of pH:
Each enzyme has a optimal pH
Too acidic or basic can disrupt ionic bonds or protonation states in the active site —> reducing activity
Prediction based on active sites
Active site containing acidic residues (like Asp or Glu) may work best at low pH
Basic residues (like His or Lys) may prefer neutral or high pH
Shape and charge of the active site depends on correct protonation
Know the differences between competitive, noncompetitive, and uncompetitive inhibitors. How do they affect V max and K M ? How do their MM and LB plots look?
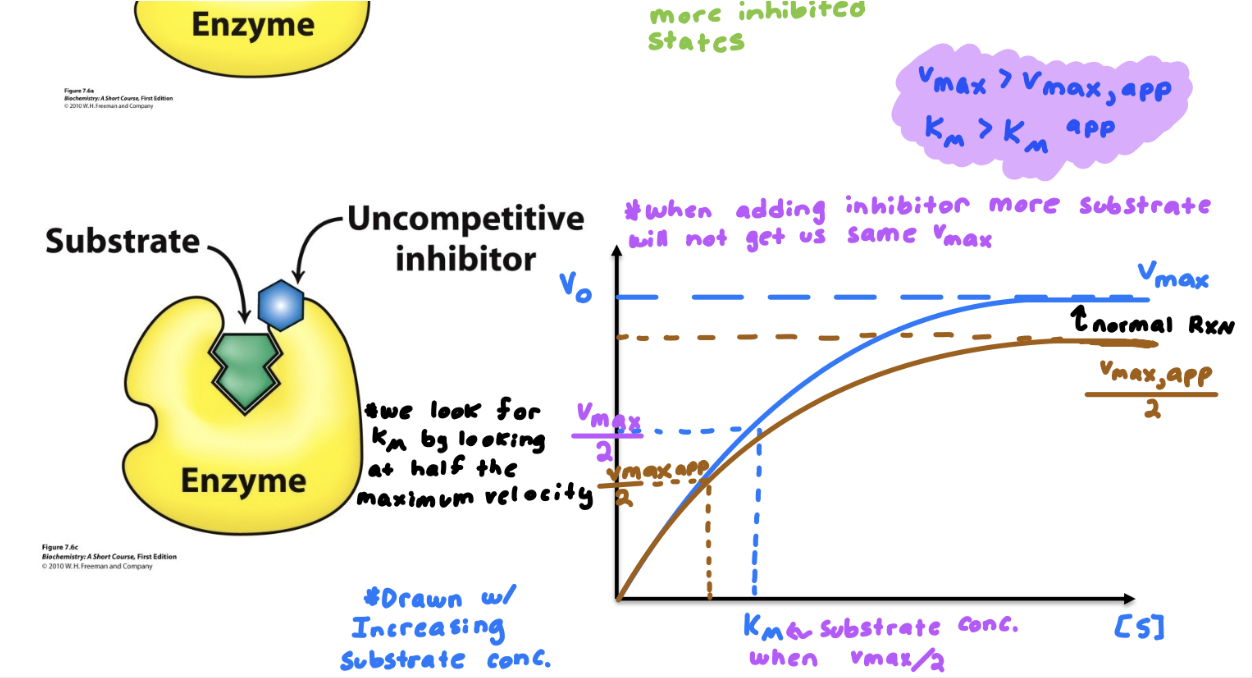
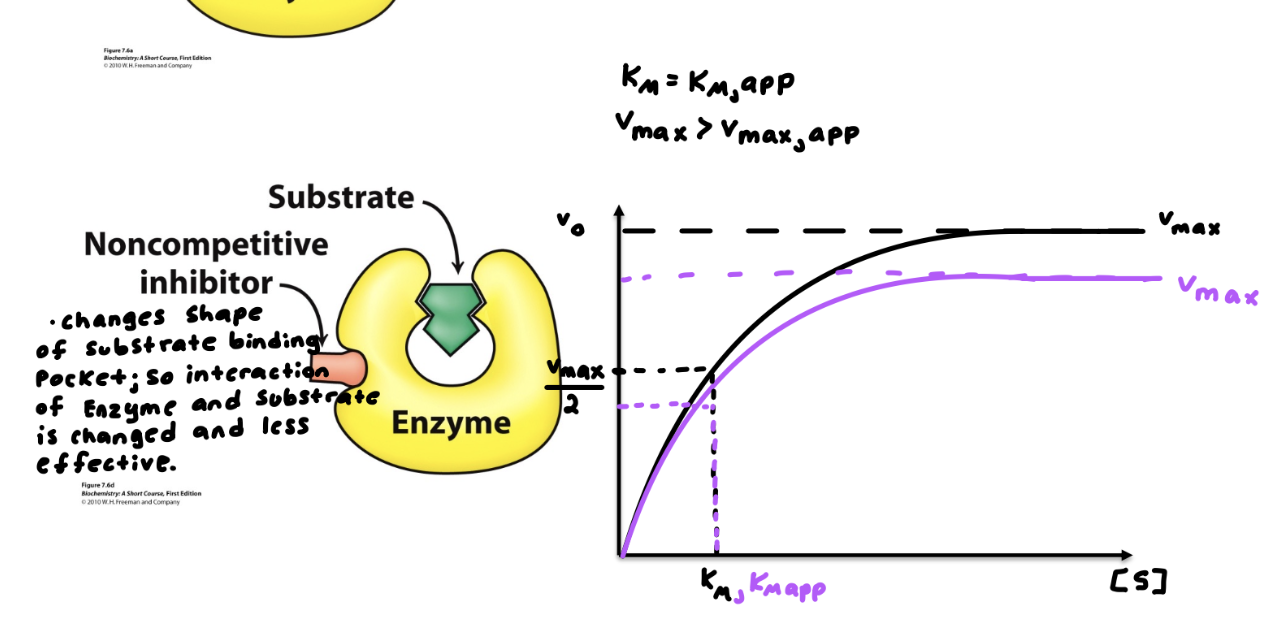
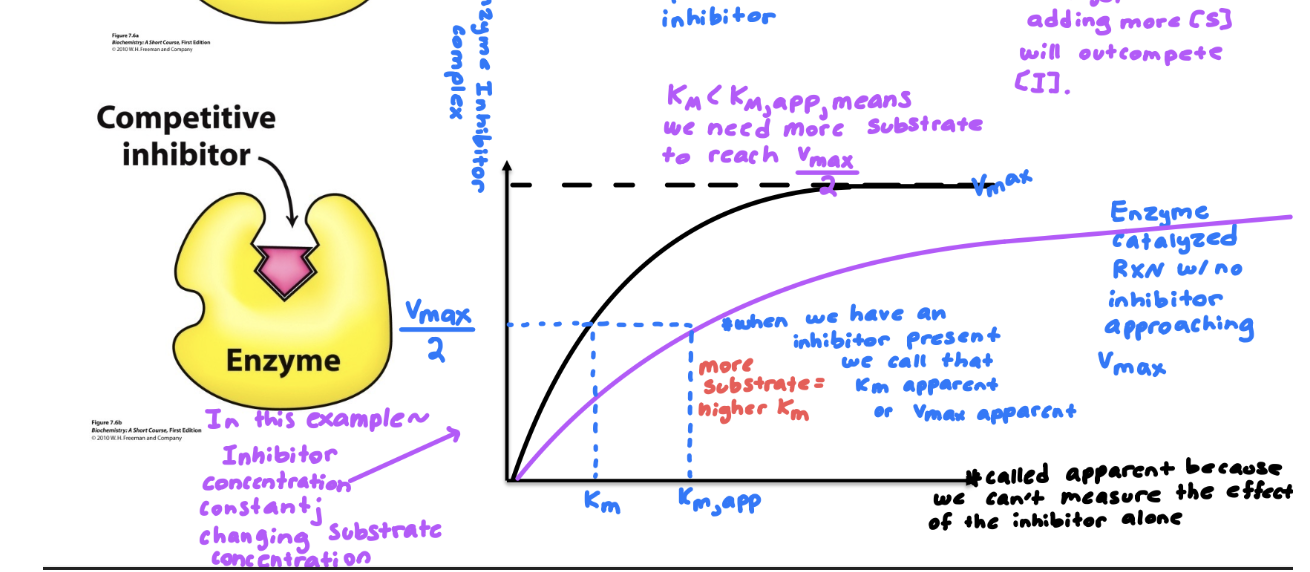
Describe examples of irreversible inhibitors
Explain how Chymotrypsin works
Summary: Whole point of this regeneration in Chymotrypsin is to allow the enzyme to be reused repeatedly-catalyzing many reactions without being consumed. Focusing on efficiency
Two step reaction
Step 1: Acylation (fast)
Acylation is the addition of R-CO- ; in this case its to Serine-195 where it becomes covalently bonded.
Aspartate-102 stabilizes the positive charge on Histidine by H-bonding to it(enhancing its ability to de-protonate)
Histidine-57 acts as the base accepting protons from Serine-195 to activate it as a nucleophile
The oxyanion Hole is found within both these phases, it ultimately forms a stabilizing pocket that stabilizes the negative charge on oxygen during the intermediate step
Summary: Substrate binds in active site, amine product is released (first product) forming acyl-enzyme intermediate
Step 2: De-acylation (slow)
In De-acylation R-CO- ; becomes removed from Serine-195 restoring the enzyme ultimately.
Aspartate-102 stabilizes the positive charge on Histidine by H-bonding to it (enhancing its ability to de-protonate)
Histidine-57 acts as the base again for water activating it as the nucleophile to attack the acyl-enzyme intermediate.
The oxyanion Hole is found within both these phases, it ultimately forms a stabilizing pocket that stabilizes the negative charge on oxygen during the intermediate step
Summary: Water enters the acyl-enzyme intermediate, carboxylic acid (second product) is released, and enzyme is re-generated
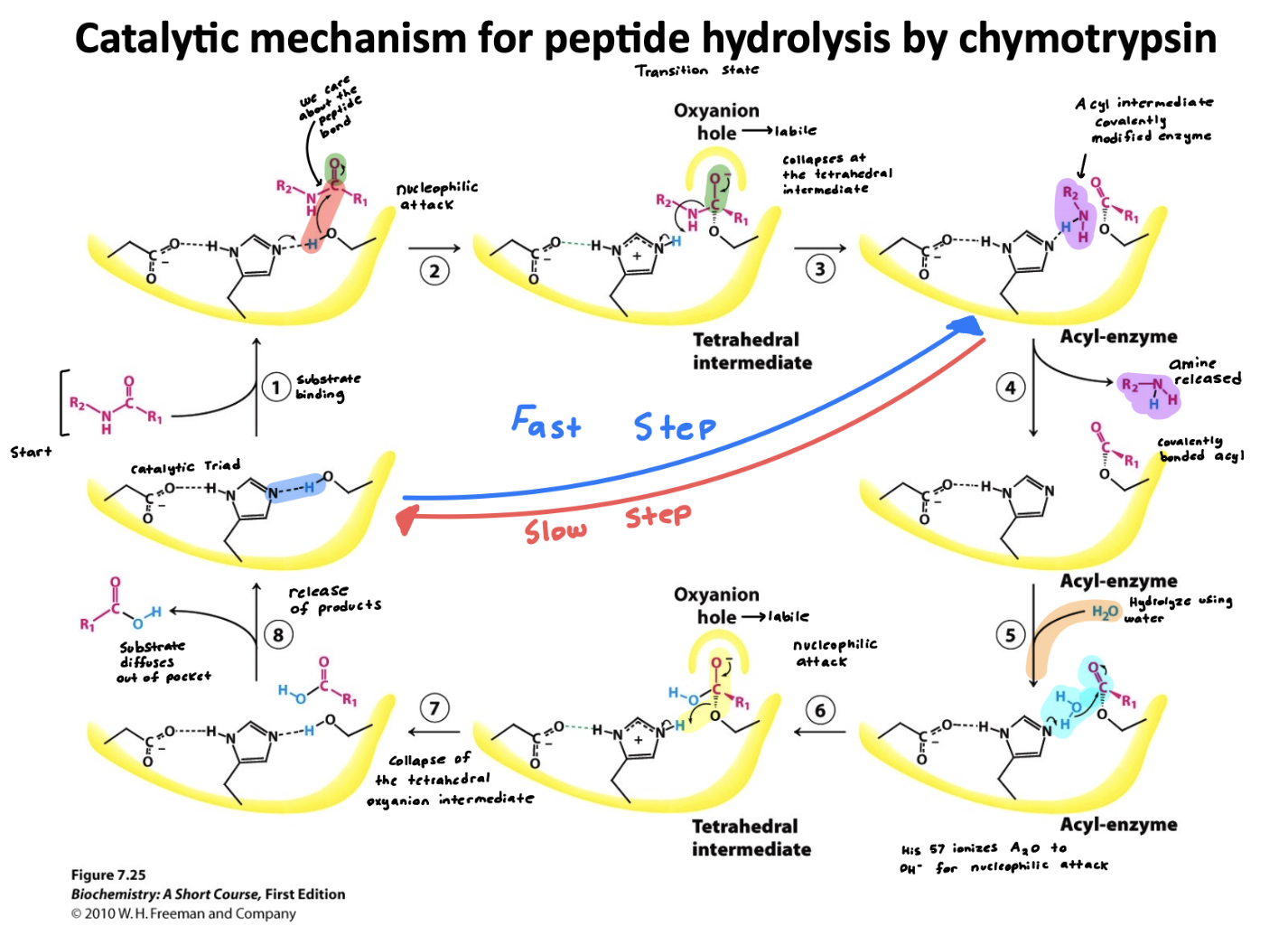
Define a catalytic triad and describe the function of each amino acid in the catalytic triad
A catalytic triad is a group of three coordinated amino acids in an enzymes active site that work together to carry out catalysis
Serine 195- acts as a nucleophile, attacking the substrates peptide bond
Histidine 57- acts as the base, accepting a proton from serine to activate it
Aspartate 102- stabilizes the charge on histidine
Define allostery
The regulation of an enzyme or protein by binding a molecules at a site other than the active site, called the allosteric site
Binding causes conformational change in the protein
This change can increase (activator) or decrease (inhibitor) enzyme activity
Common in enzymes with multiple subunits like hemoglobin
Describe the two models of allostery
Concerted model-
All subunits of the enzyme exist in either low-affinity (tense, T) state or a high affinity (relaxed, R) state
Enzyme switches between these states as a whole unit- all or none
Features: Binding of substrate shifts the equilibrium from the T state to the R state.
Sequential model-
subunits change individually upon substrate binding
binding of substrate to one subunit induces a conformation change in that subunit, where the change is then passed to neighboring subunits, increasing their affinity.
Features: not all subunits have to be in the same state
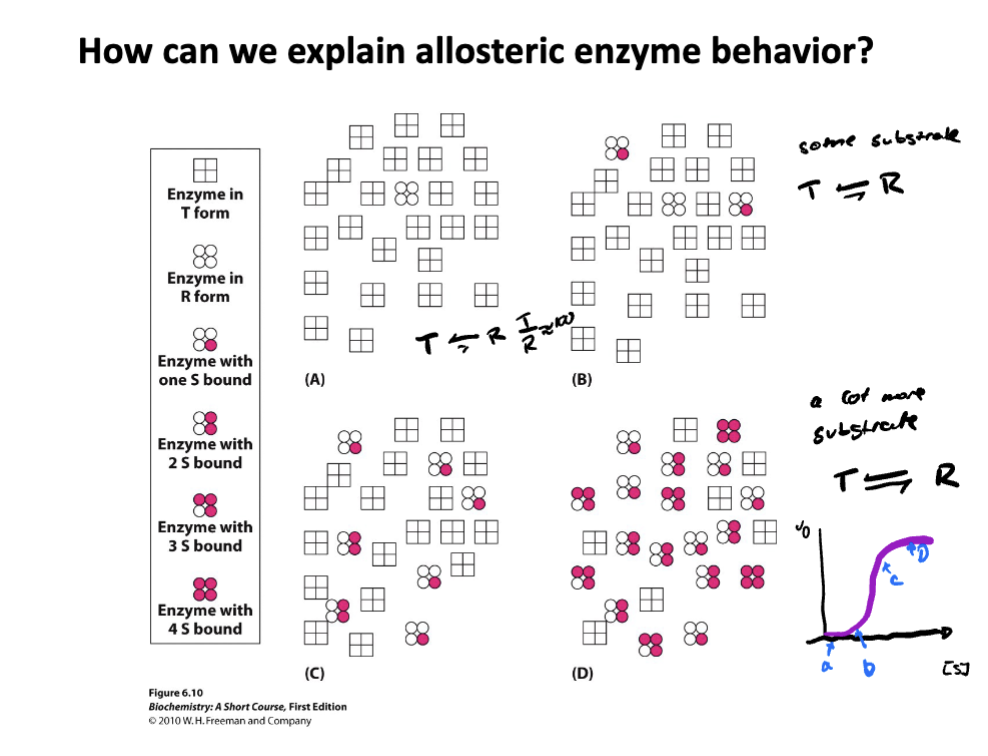
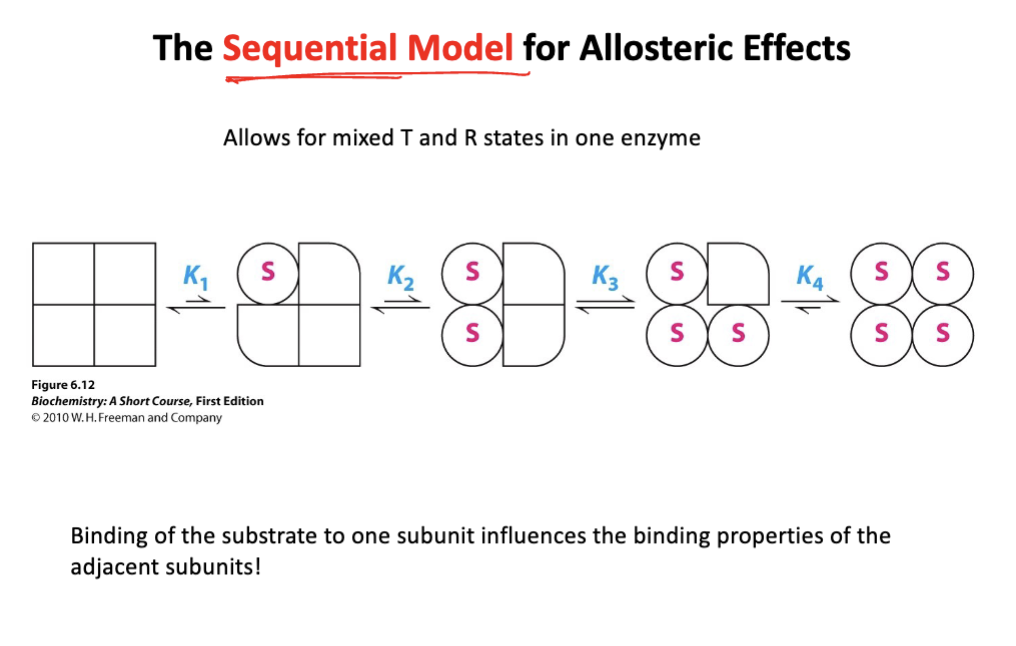
Describe how effectors regulate hemoglobin
Oxygen: A positive (homotropic) effector (activator)
Binding of oxygen increases the affinity for more oxygen (cooperative binding)
shift hemoglobin between the T (tense state, low-affinity) and R(relaxed, high affinity) states, fine tuning oxygen delivery based on the body’s needs
2,3-BPG: A negative (heterotrophic) effector (inhibitor)
Binds to the center of hemoglobin and reduces its affinity for oxygen, helping release oxygen in tissues.
Increases and stabilizes the T state of hemoglobin
Define isomer, constitutional isomer, epimer, stereoisomer, diastereoisomer, and anomer
Isomer: Have the same molecular formula but have different structures (branches constitutional Isomers and Stereoisomers)
Constitutional Isomers: Differ in the order of attachment of atoms
Stereoisomers: Atoms are connected in the same order but differ in spatial arrangement
Enantiomers: non-superimposable super image (A type of stereoisomer)
Diastereoisomer: Isomers that are not mirror images
Epimers: Differ at one of several asymmetric carbon atoms
Anomers: Isomers that differ at a new asymmetric carbon atom formed on ring closure.
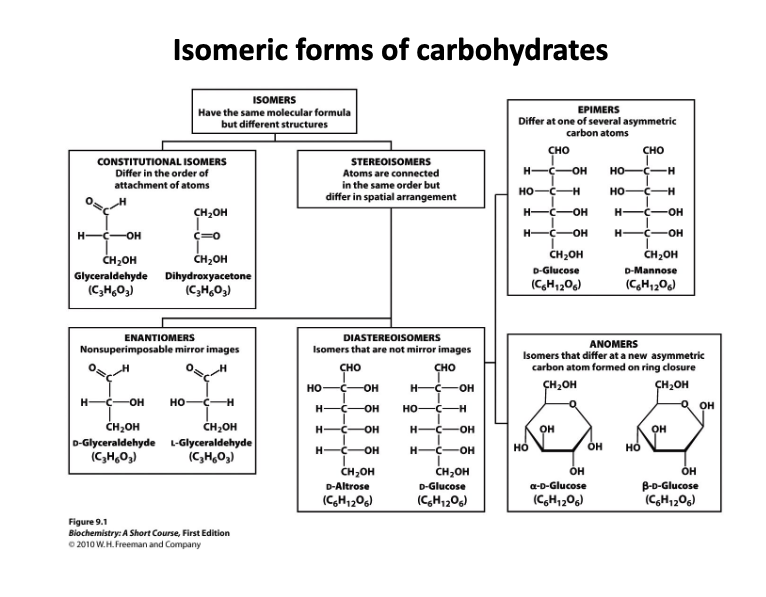
Draw glucose, mannose, and galactose in the Fisher representation

Describe storage forms and structural forms of carbohydrates
Storage forms of carbohydrates
Glycogen (animals): Highly branched Alpha (1 —>4) and Alpha (1 —>6) glucose polymer. stores in liver and muscle
Starch (plants): mixture of amylose (linear Alpha (1 —>4) and amylopectin (branched,Alpha (1 —>6). Plant energy storage
Structural forms of carbohydrates
Cellulose (plants): beta (1—>4) linked glucose. Forms rigid fibers; found in plant cell walls
Chitin (fungi, insects): Beta (1—>4) linked in N-acetlyglucosamine. Tough structure of exoskeletons
Describe and recognize different forms of glycosidic bonds
Alpha (1 —>4): is found in starch and glycogen
Beta (1 —>4): Found in cellulose, creates a straight, rigid structure
Alpha (1 —>6): Causes branching in glycogen and amylopectin]
Alpha is going down (like fish) (axial)
Beta is going up (equatorial)
Why are storage forms branched?
Storage forms like Glycogen and starch are branched
Branching creates many ends, where enzymes can simultaneously break down glucose
Branched structures are compact and soluble, allowing high-density storage of glucose in small spaces
Overall: Branching= Faster access + Efficient packing
How do carbohydrates relate to human blood types
Blood type is determined by the type of carbohydrate (sugar) attached to the cell surface proteins on red blood cells
Describe the structure and function of glycoproteins, proteoglycans, and glycosaminoglycans
Structure
Glycoprotein: protein + smaller glycosylation (short carbohydrate chain)
Proteoglycan: mostly carbohydrate + smaller protein/peptide as the scaffold
Glycosaminoglycans: long unbranched chain of disaccharides (long sugar chain)
Function
Glycoprotein: involved in cell signaling, immune response, and cell recognition
Proteoglycan: Structural support and cushioning (cartilage)
Glycosaminoglycans: Maintains extracellular matrix structure, traps water
Know the five classes of lipids and their chemical properties
Free fatty acids (FA)- Fuel, important building block of membrane lipids
Triacyl glycerides- Storage form of FA
Phospholopids- FA attached to phosphate head group
Glycolipid- Carbohydrate attached to lipid
Steroid- Polycyclic hydrocarbons; (harmones, components of membranes)
Know fatty acid numbering and nomenclature. Be able to draw a lipid according to nomenclature
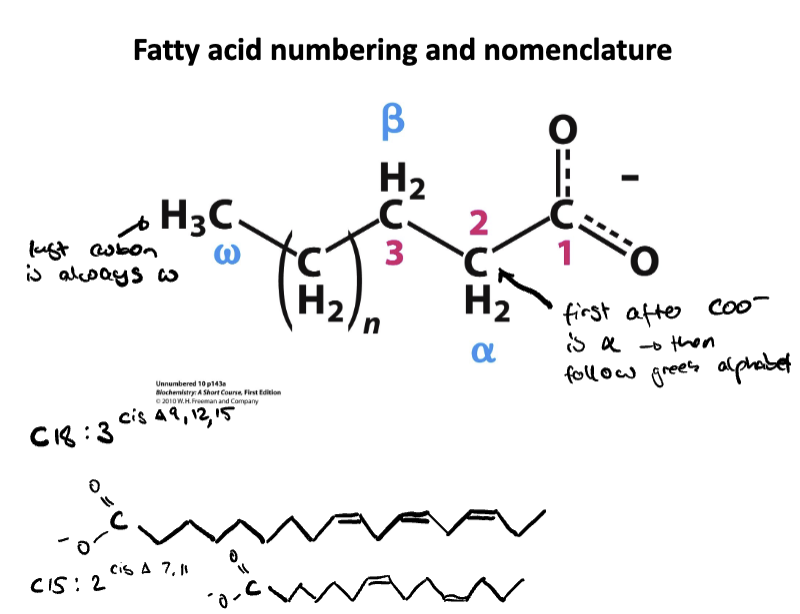
Explain why unsaturated fatty acids are important for health
Support cell membrane fluidity
Provide essential fats (like omega-3) that the body cannot make
Improve heart health by lowering bad cholesterol
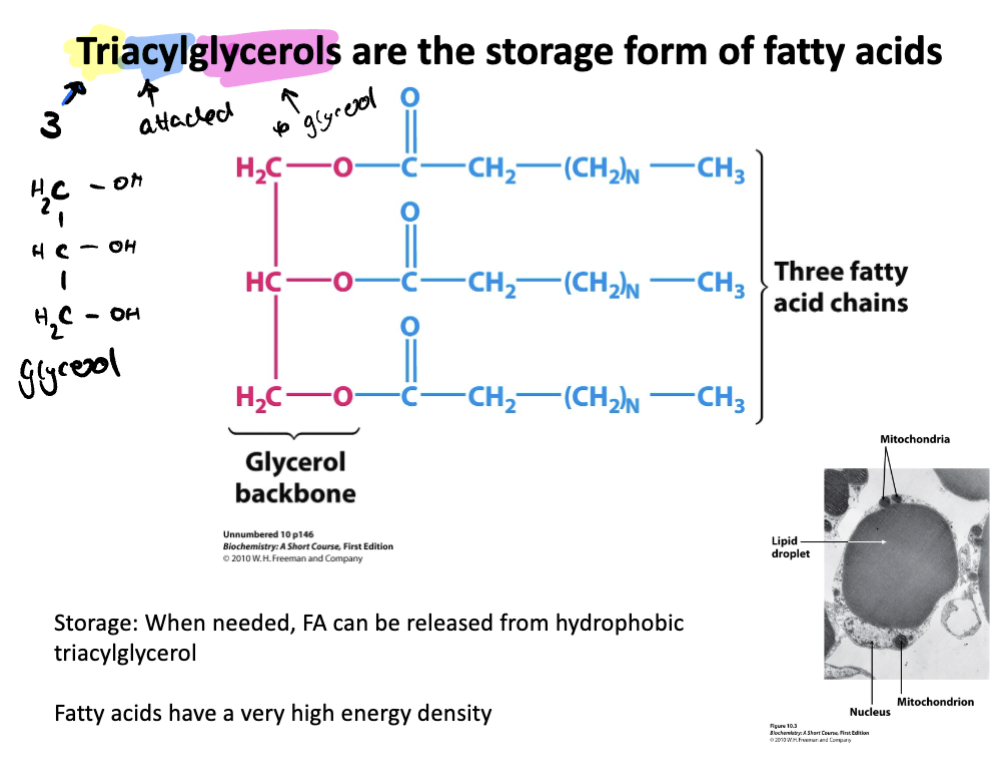
Describe the storage form of fatty acids
Triacylglycerols (3 attached to glycerol) are the storage form of fatty acids
made up of glycerol backbone attached to three fatty acid chains
Describe the structure and function of DNA polymerase
Structure shaped like a hand with three domains
palm: Catalytic site (where DNA synthesis occurs)
Fingers: positions incoming nucleotides
Thumb: holds and stabilizes the DNA
Function
Adds nucleotides to the 3 prime end of a growing DNA strand using a template strand
Proofreads some DNA polymerases that have 3 to 5 prime exonuclease activity
How does DNA polymerase proofread?
DNA polymerase proofreads by switching to its own exonuclease active site where it allows it to bind then cleaves nucleic acid if wrong to remove mismatched bases
Backtracks after removal and gets another try at added the correct dNTP
Describe the role of each enzyme involved in DNA replication
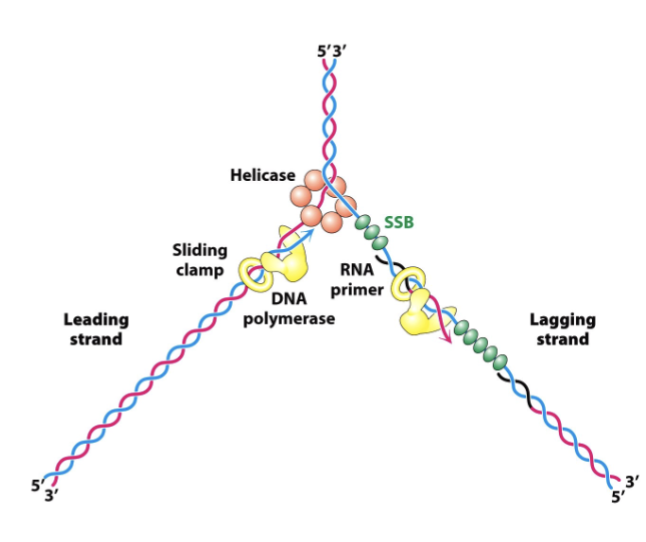
Helicase: unwinds the DNA double helix
SSBs: Stabelizes unwound DNA
Primase: adds RNA primers
DNA polymerase: adds nucleotides 5’ to 3’ using the template strand
Leading strand: synthesized continuously
lagging strand: discontinuously synthesized in Okazaki fragments
DNA polymerase: replaces primers with DNA
Ligase: seals DNA fragments
Topoisomerase: relieves tension ahead of the fork
Sliding clamp: prevents DNA polymerase from falling off the DNA template strand during replication
RNA primer: Provide starting point for DNA synthesis by offering a free 3’ OH group for DNA polymerase to being adding DNA nucleotides
Describe how DNA replication is carried out on the molecular level
Helicase unwinds the DNA double helix
Single-strand binding proteins (SSBs) stablize the unwound strands
Primase lays down the RNA primers
DNA polymerase adds nucleodies synthesizing 5’ to 3’
Leading strand is made continuously
lagging strand is made in okazaki fragments that is sealed by DNA ligase
DNA polymerase proofs reads and corrects mismatched bases using its 3’ to 5’ exonuclease activity
Ultimately Replication is semi-conservative, producing two DNA molecules, each with one old strand and one new strand
What are telomeres and telomerase and why do we need them?
Telomeres: repetitive DNA sequences at the ends of chromosomes that protect them from damage
Telomerase: An enzyme that adds more telomere repeats to the ends of DNA
why doe we need them?
We need telomeres because each time DNA replicates, the ends get shorter
We need telomerase because it prevents important genes from being lost by extending telomeres, especially in stem cells and gene cells (ultimately maintains them to prevent gene loss during DNA replication)
Describe the trombone model
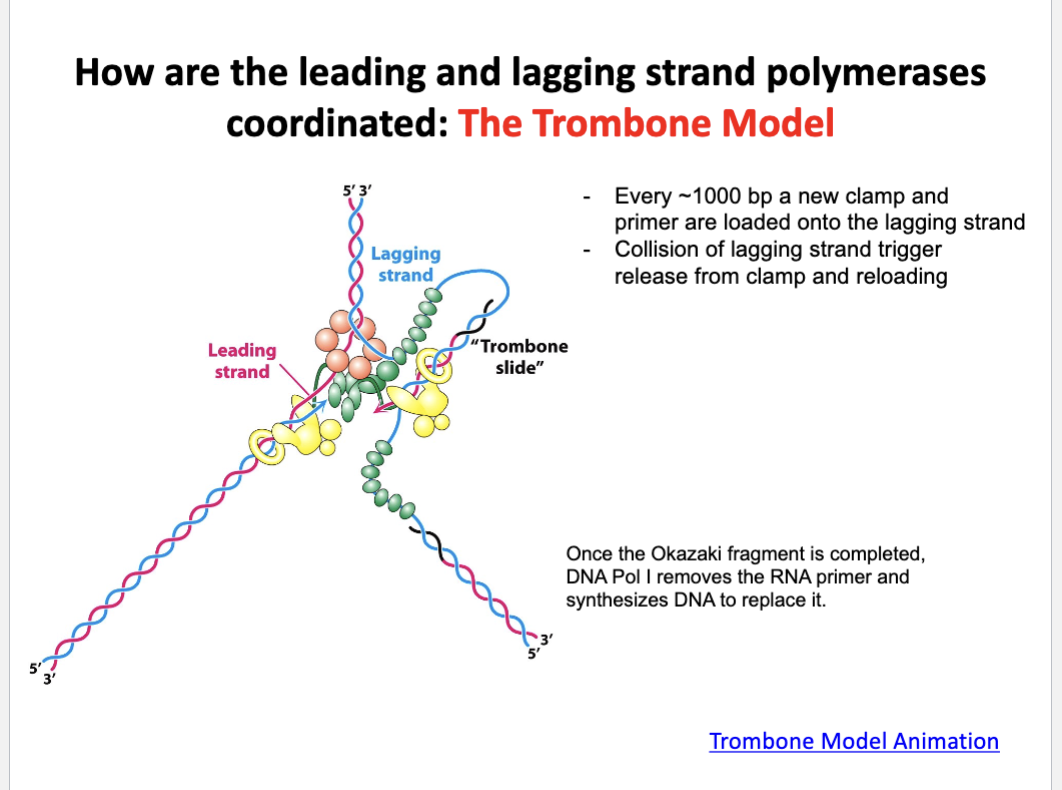
The trombone model explains how the leading and the lagging strands are synthesized simultaneously
The lagging strand loops out like a trombone slide allowing synthesis in the same direction as the fork
each time an Okazaki fragment is finished, the loop resets with a new primer and clamp
this keeps both polymerases moving together despite opposite strand orientation
Describe the role of each enzyme involved in RNA transcription.
sigma factor: Helps RNA polymerase find and bind to the promoter region to start transcription
RNA polymerase: Main enzyme that synthesizes RNA
Helicase: Unwinds the DNA exposing the template strand
Topoisomerase: relieves supercoiling ahead of the bubble to prevent DNA tangling
Compare and contrast similarities and differences between RNA transcription and DNA replication
Both
Use DNA template
Both build new strand in the 5’ to 3’ direction
DNA replication
Uses Thymine as base
Uses both template strands
has proof reading
RNA transcription
Uses Uracil as base
Uses one template strand
no proofreading (has more mistakes)
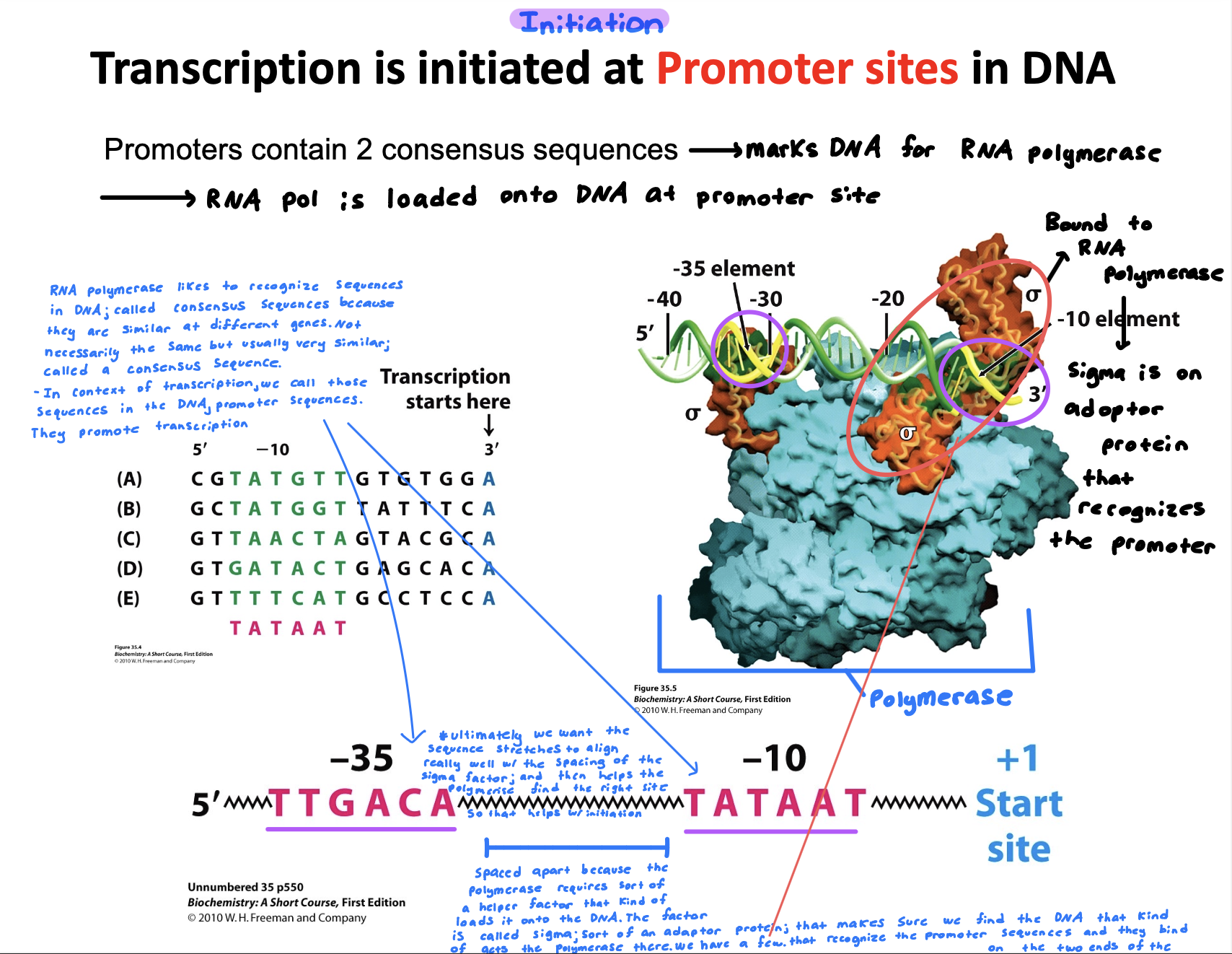
Describe how transcription is initiated
Transcription is initiated when RNA polymerase binds to a specific promoter sequence (promoters have 2 consensus sequence on the DNA
Sigma factors (2 areas) bound to the RNA polymerase recognize the promoter sequence
Ultimately we want the sequence stretch to align really well with the spacing of the sigma factor which then helps the polymerase find the rise site in return helping initiation
Extra info:
RNA polymerase likes to recognize sequences in DNA called consensus sequences because they are similar at different genes. This is referred to as promoter sequences.
The sequence is spaced apart because the polymerase requires sort of a helper factor that kinds of loads it into the DNA. The factor is called sigma sort of an adaptor protein that makes sure we find the DNA that kind of acts like the polymerase there. We have. few that recognize the promoter sequence and they bind on the two ends of the polymerase
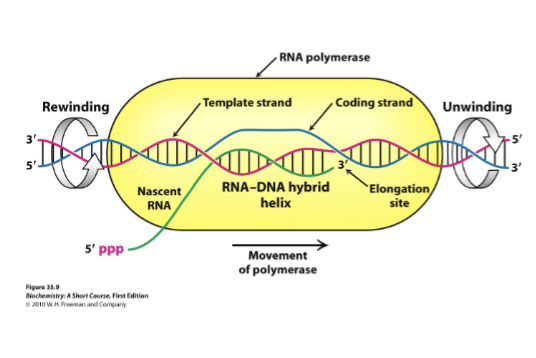
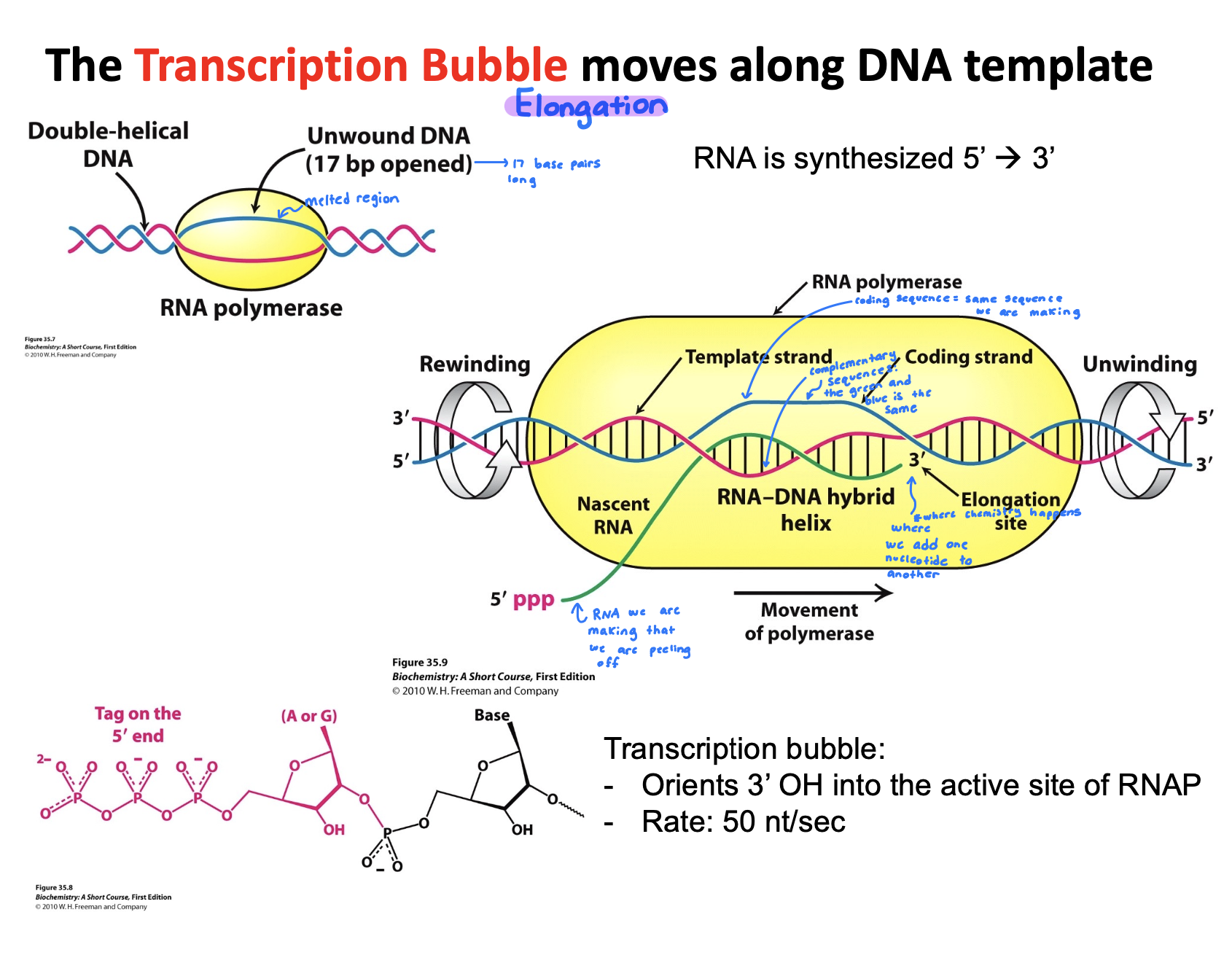
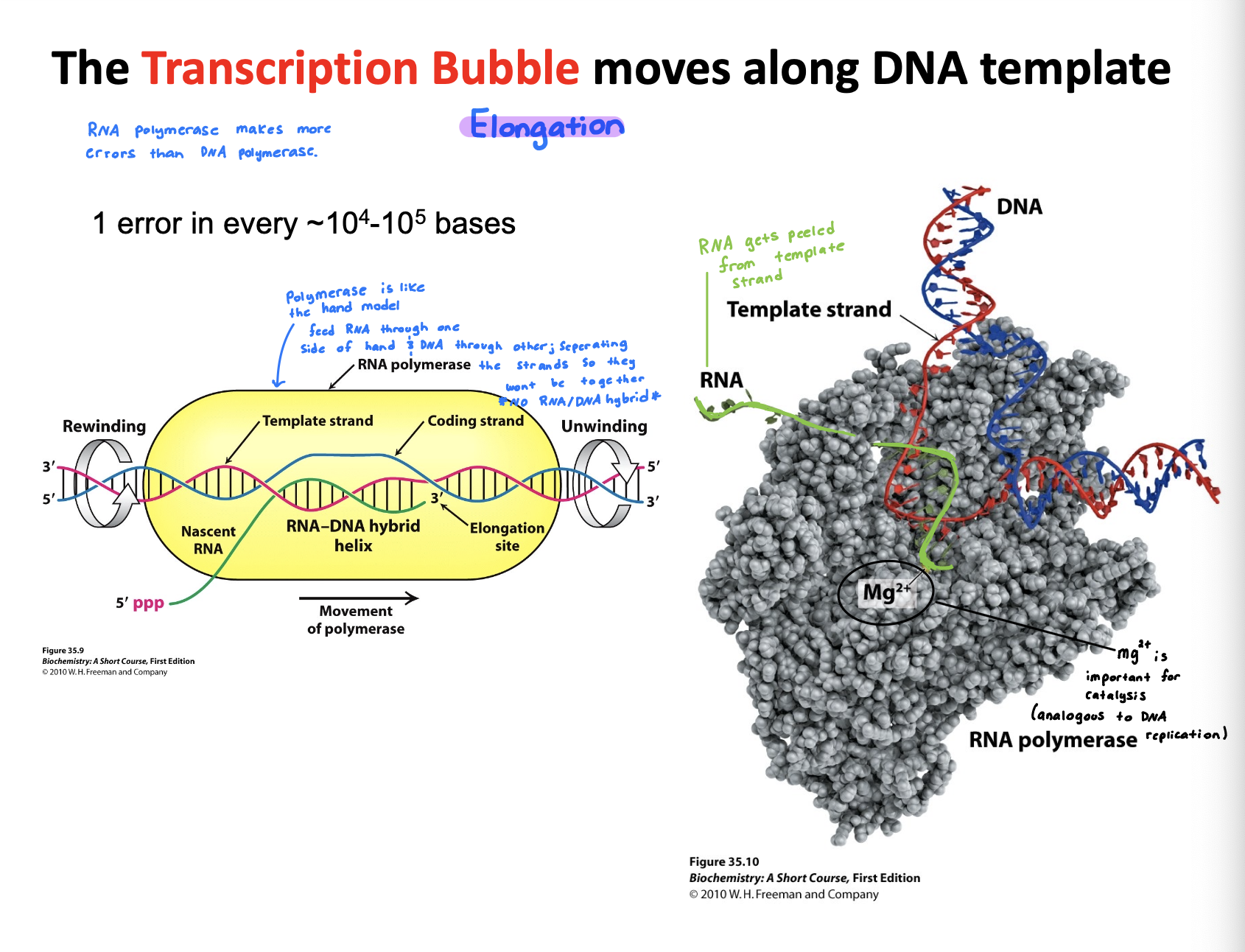
Describe the events taking place in a transcription bubble.
Elongation ultimately takes place in the transcription bubble
Nascent RNA: Newley synthesized RNA strand grows that is exiting through polymerase as the bubble shifts
Template strand: DNA strand that RNA polymerase reads to make complementary DNA
Coding strand: Serves as reference strand whose sequence is identical to the RNA transcript, helping identify gene sequence
Elongation site: Active site where RNA strand grows, using template strand to guide nucleotide addition
DNA rewinds as RNA polymerase moves forwards, the DNA re-anneals
Extra info:
RNA polymerase makes more errors than DNA polymerase
Different modes of Transcription Termination?
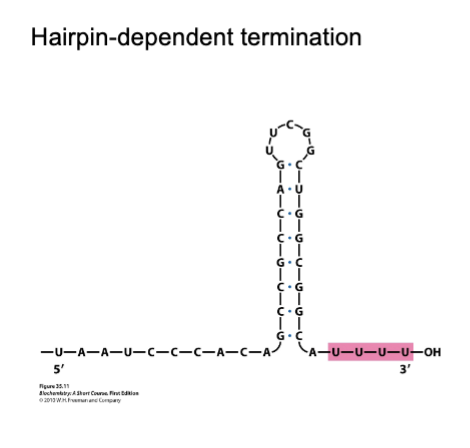
Hairpin-dependent termination
Formation of hairpin at the exit tunnel inducing stalling of RNAP
Poly-U forms weak hybrid in the active site, which leads to dissociation
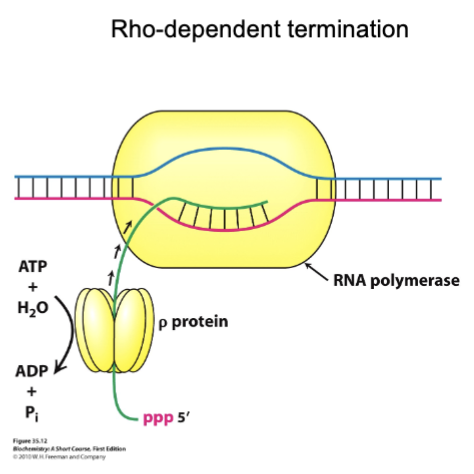
Rho-dependent termination
Mechanical process that physically strips RNA out of the transcription bubble (requires ATP)
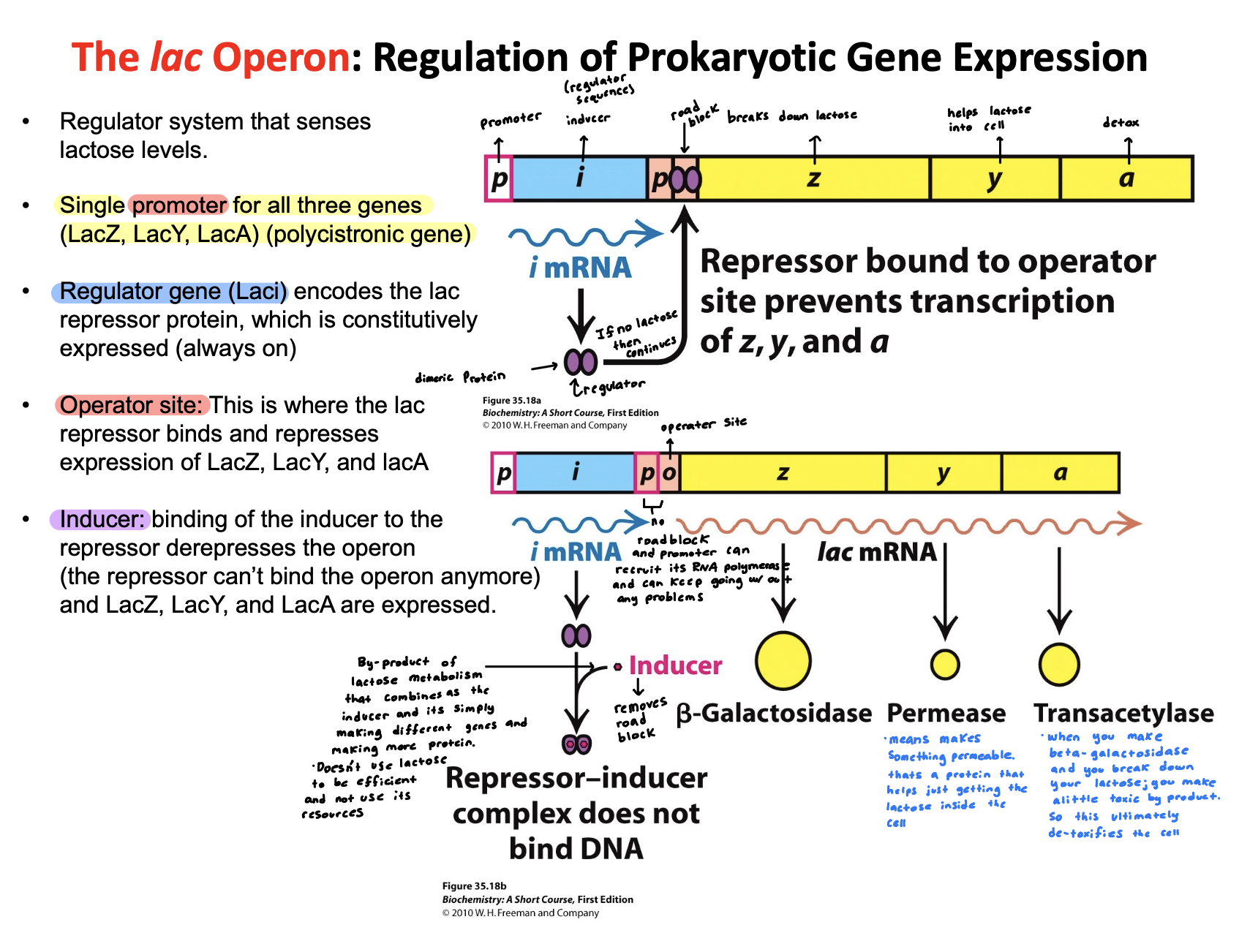
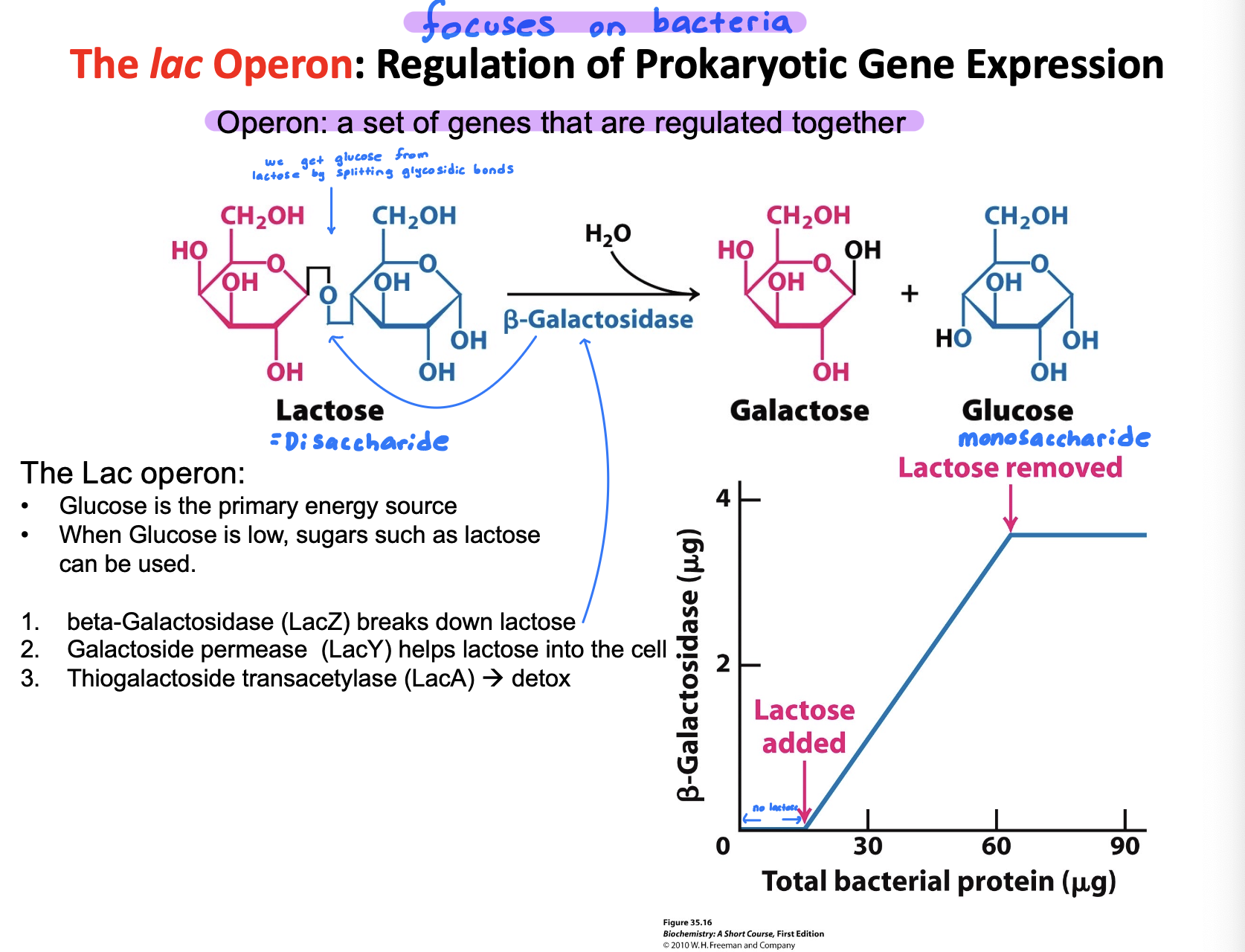
Describe how the lac operon regulates gene expression
Lac operon regulates gene expression by turning lactose-metabolism genes on or off based on nutrient ability.
No lactose: A repressor binds to the operator—> genes off
Lactose present: Lactose side product binds the repressor inducing it —→ repressor releases —→ genes on
This ensures the operon is only active when lactose is available and glucose is low
Explain why we have 64 codons but only 20 amino acids
We have 64 codons but only 20 amino acids because genetic code is degenerate meaning multiple codons can code for the same amino acid
(Alternatively) We only have 20 amino acids, but we need 64 different codons because some amino acids get more than one codon. 3 nucleotides together are one codon because they encode for one amino acid
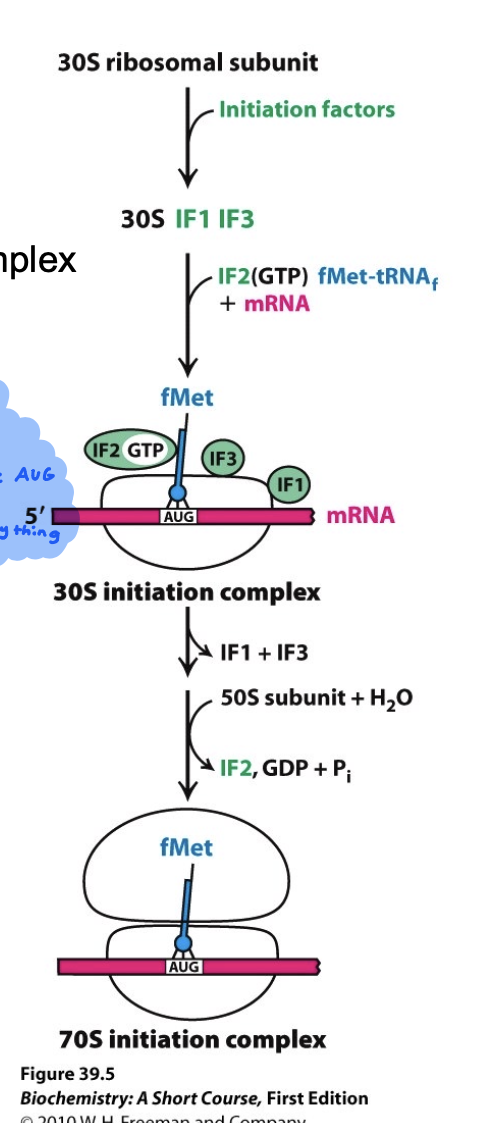
How does translation initiation work in bacteria?
30S subunit binds to mRNA at the Shine Dalgarno sequence
Shine Dalgarno site positions the mRNA sequence so that AUG (start) codon is in the P-site of the ribosome
Initiator tRNA (fMet) binds the start codon (AUG)
Initiation factors (IF-1, IF-2, and IF-3) help assemble the complex
50S subunit joins, GTP is hydrolyzed, and factors are released
Ultimately the 70S initiation complex is formed and is ready for elongation
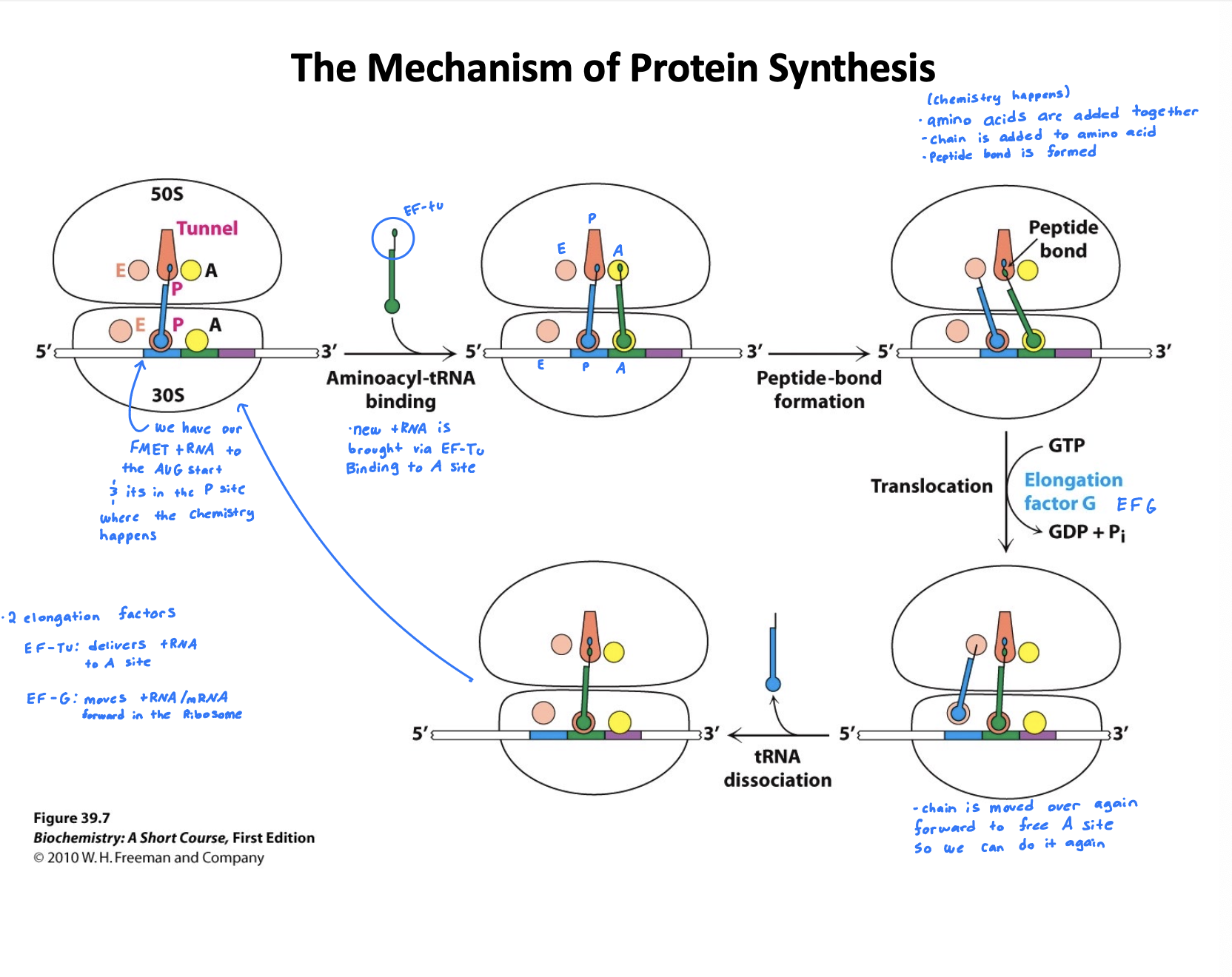
How does translation elongation work in bacteria?
Codon recognition: A charged tRNA (aminoacyl-tRNA) escorted by elongation factor EF-TU and GTP enters the A site of the ribosome. GTP is hydrolyzed and EF-Tu is released
Peptide bond formation: Chain is added to amino acid, peptide bond is formed. The growing peptide chain is transferred from the tRNA in the P site to the amino acid on the tRNA in the A site
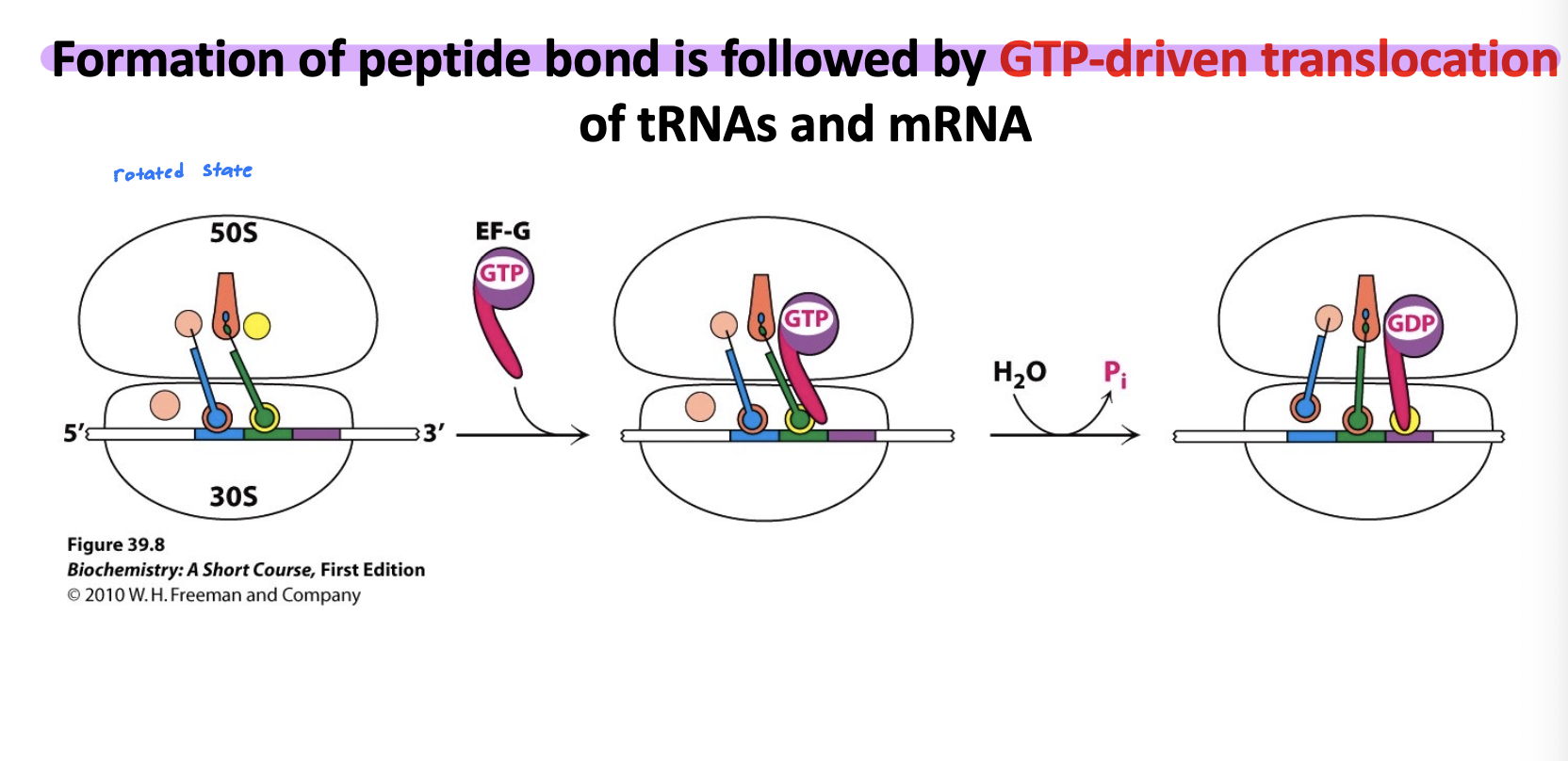
Translocation: EF-G and GTP help shift the ribosome forward by one codon. The tRNA in the P site moves to the E site, and the A-site tRNA carrying the peptide chain moves to the P site. Leaving the A site open for the next tRNA.
This whole cycle repeats until the stop codon is encountered, signaling termination
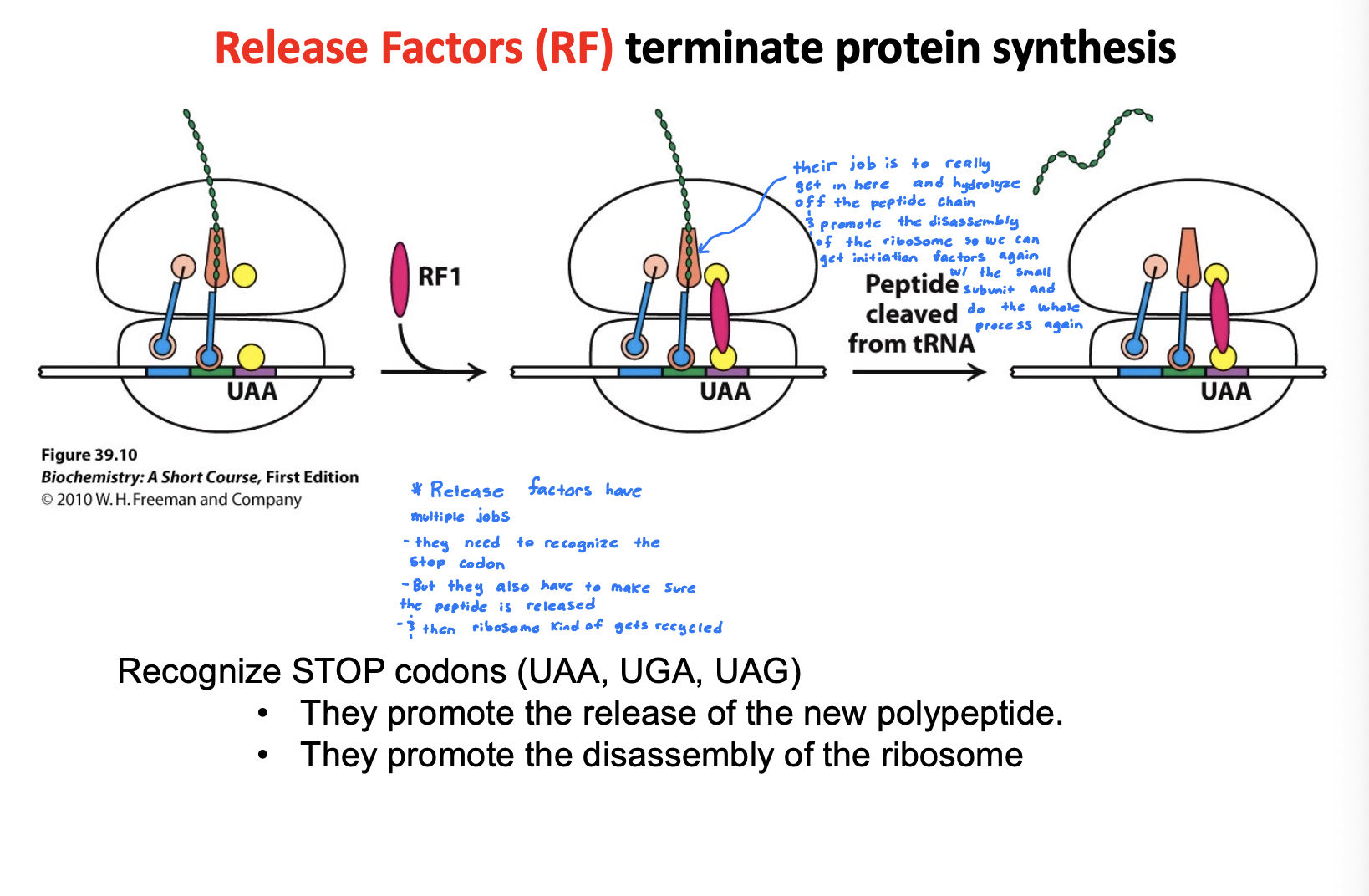
How does translation termination work in bacteria?
Release Factors (RF) terminate the protein synthesis
RF have multiple jobs, but their main job is to really get in there and hydrolyze the peptide chain off and promote the disassembly of the ribosome so we can get initiation factors again with the small subunit and do the whole process again
Generic answer:
Recognize STOP codons. They promote the release of new polypeptide. They promote the disassembly of the ribosome