Biology 305: DNA Mutations & Repair (16.1-16.4)
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
consequences of point mutations within genes
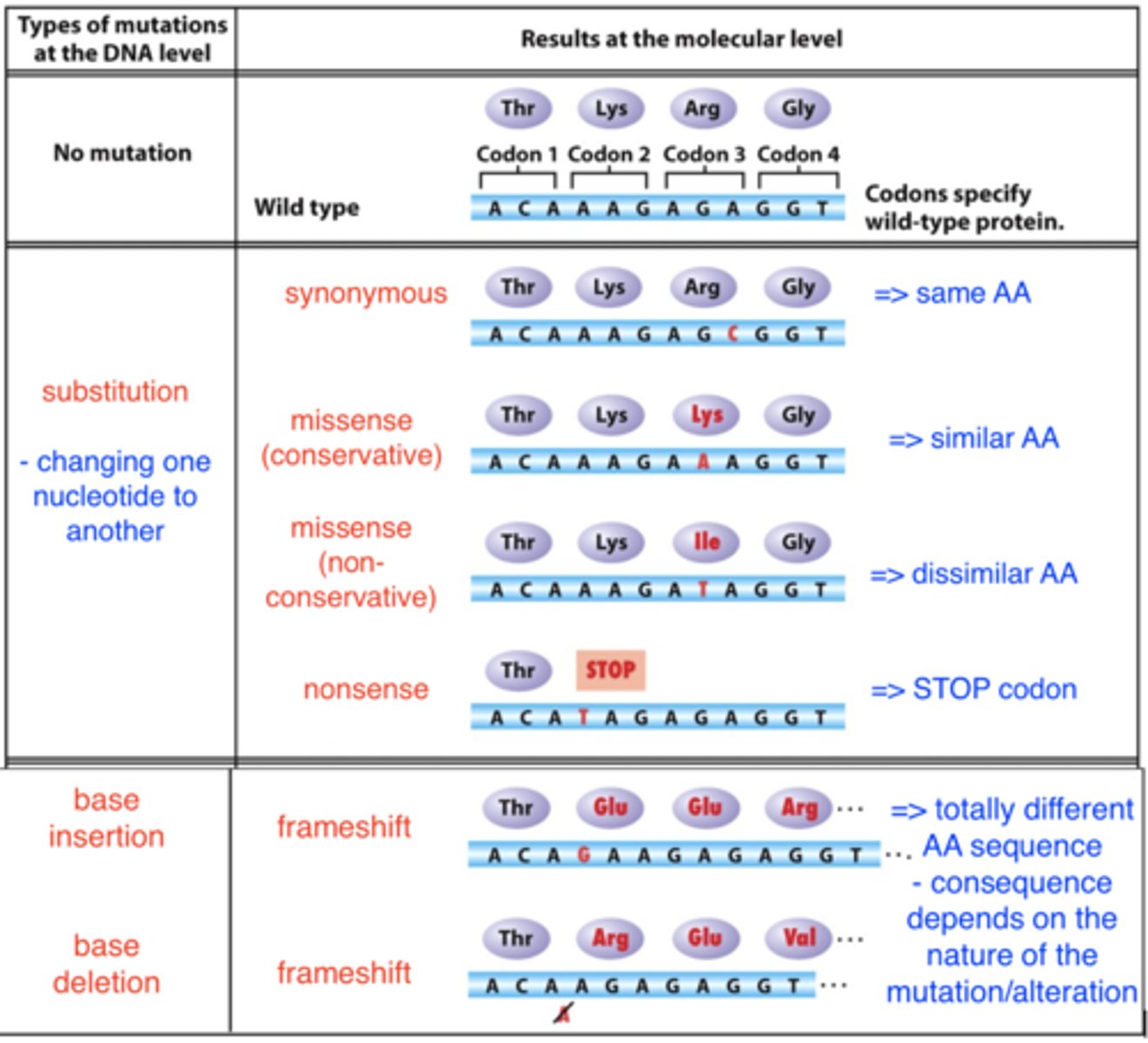
consequences of point mutations on gene products
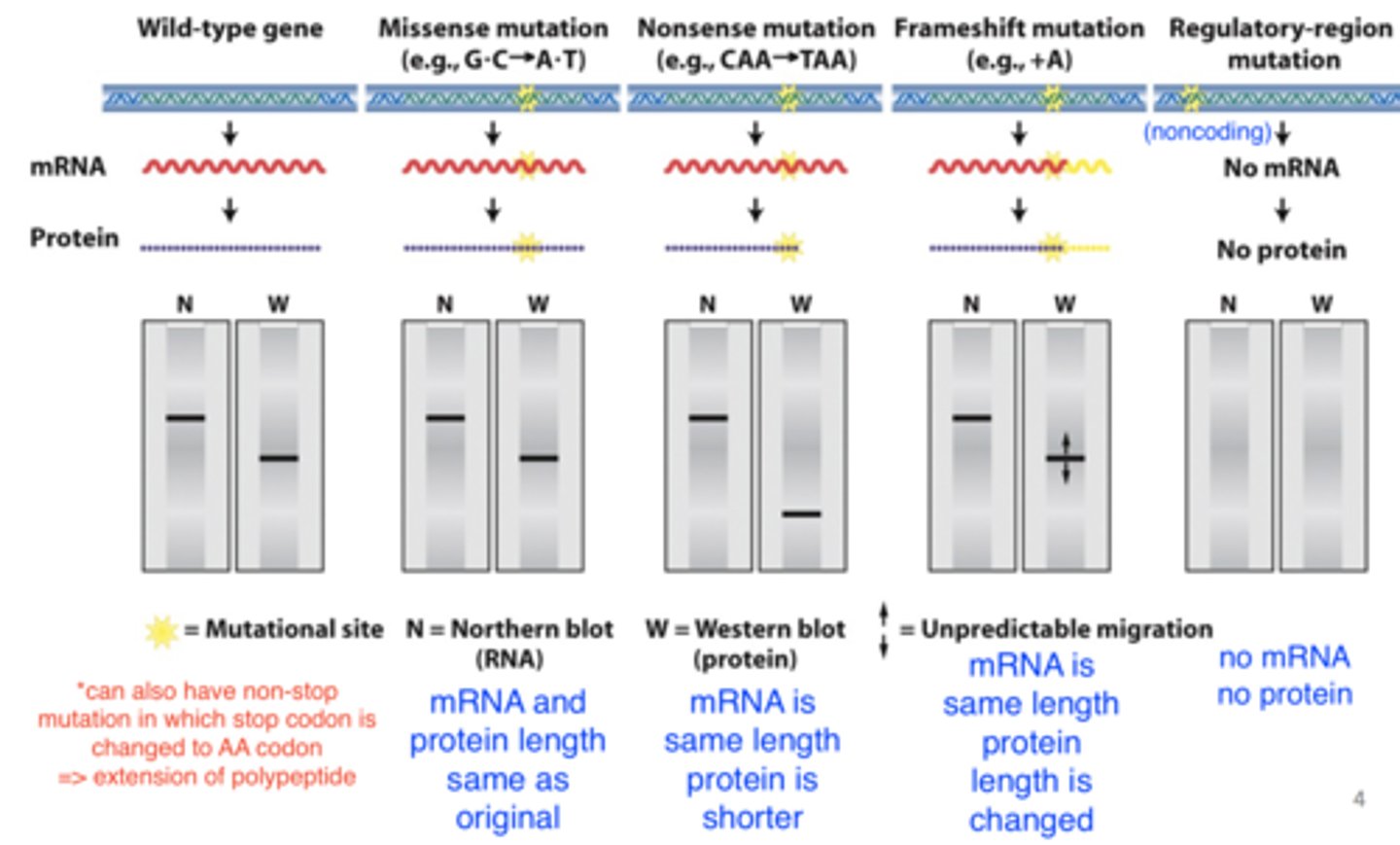
point mutations can alter ________________
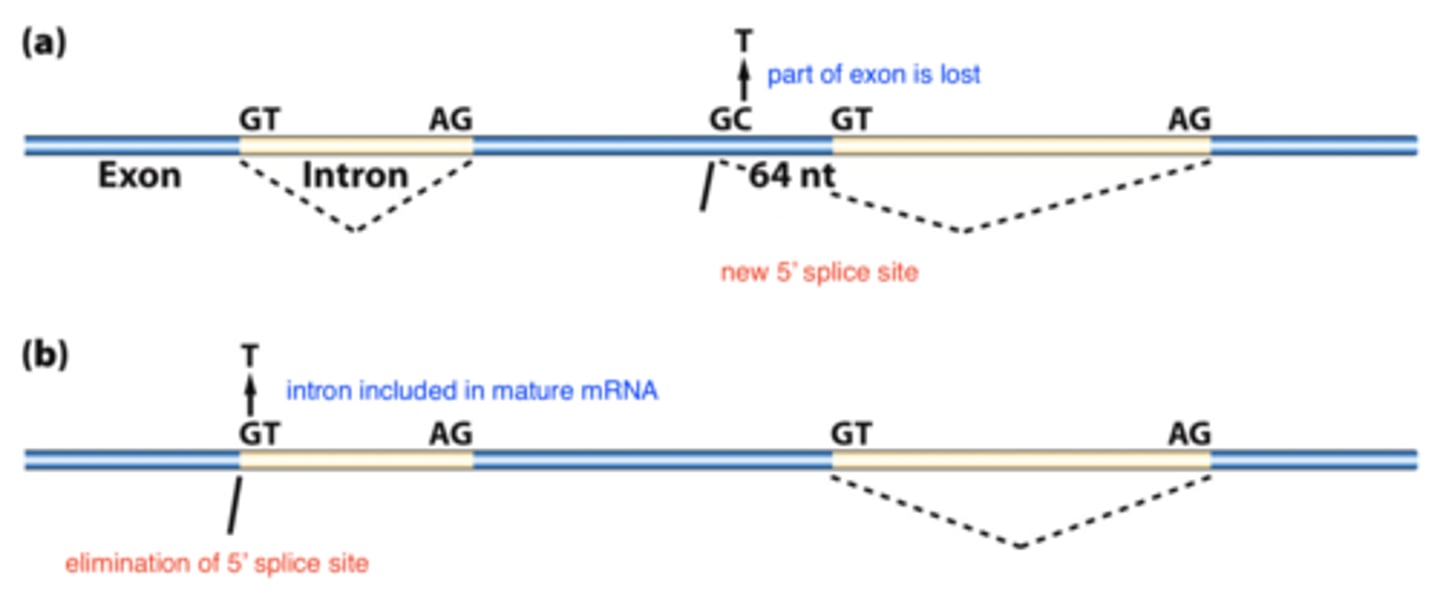
mRNA splicing
- can form new 5' slice donor site
-- mRNA shorter
-- protein length changed
- can eliminate 5' splice site
-- mRNA longer
-- protein length changed
spontaneous mutations occur due to ________________ including...
errors in DNA replication
classified based on change that is occurring:
- transitions:
-- purine => purine (ex. G-C => A-T)
-- pyrimidine => pyrimidine
- transversions:
-- purine => pyrimidine (ex G-C => T-A)
-- pyrimidine => purine
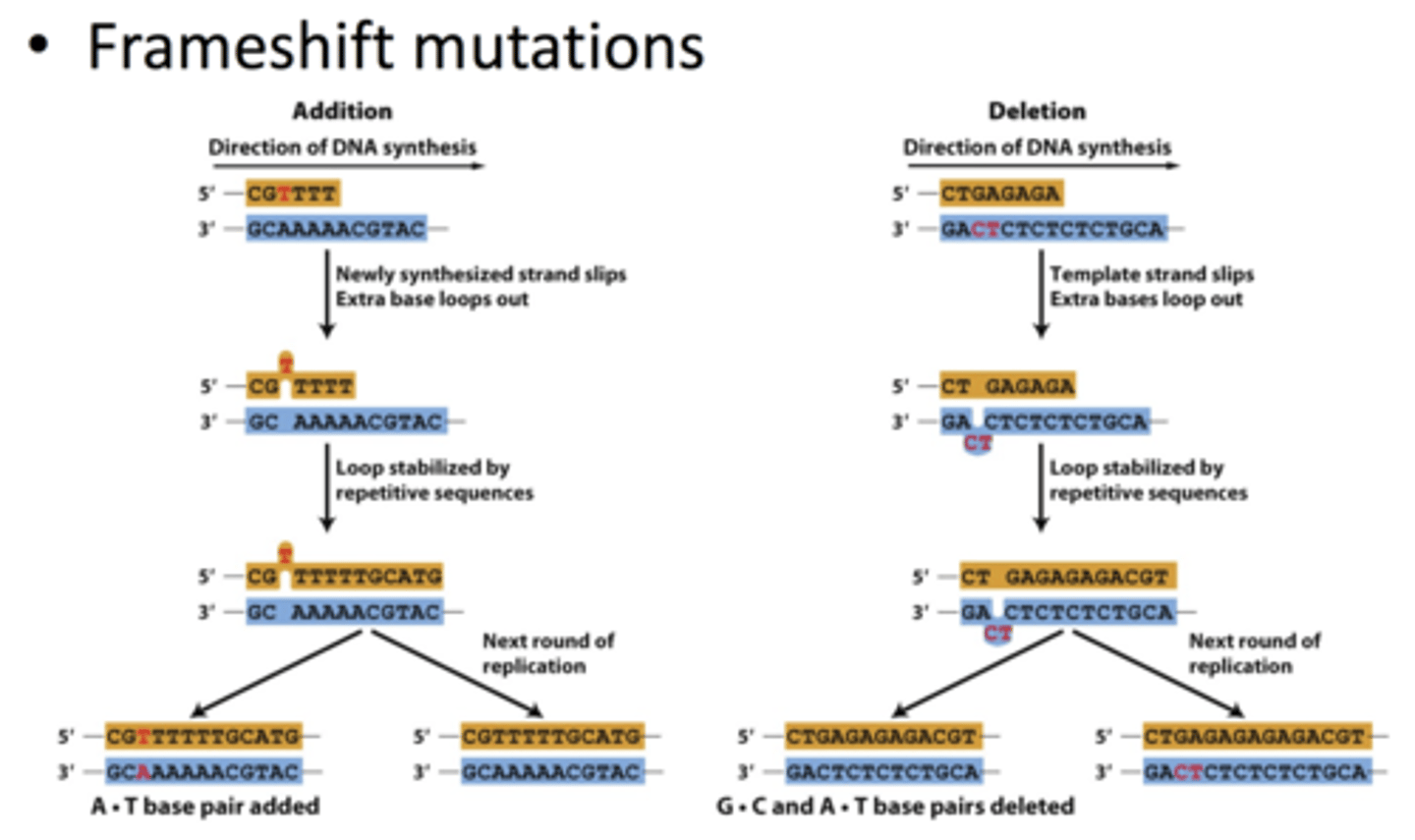
- frameshift mutations: caused by additions or deletions
-- often occur in repetitive DNA sequences
-- during replication, strand slips and offsets other strand by 1
=> extra base loops out
naturally occurring DNA damage can generate __________________ including...
spontaneous lesions
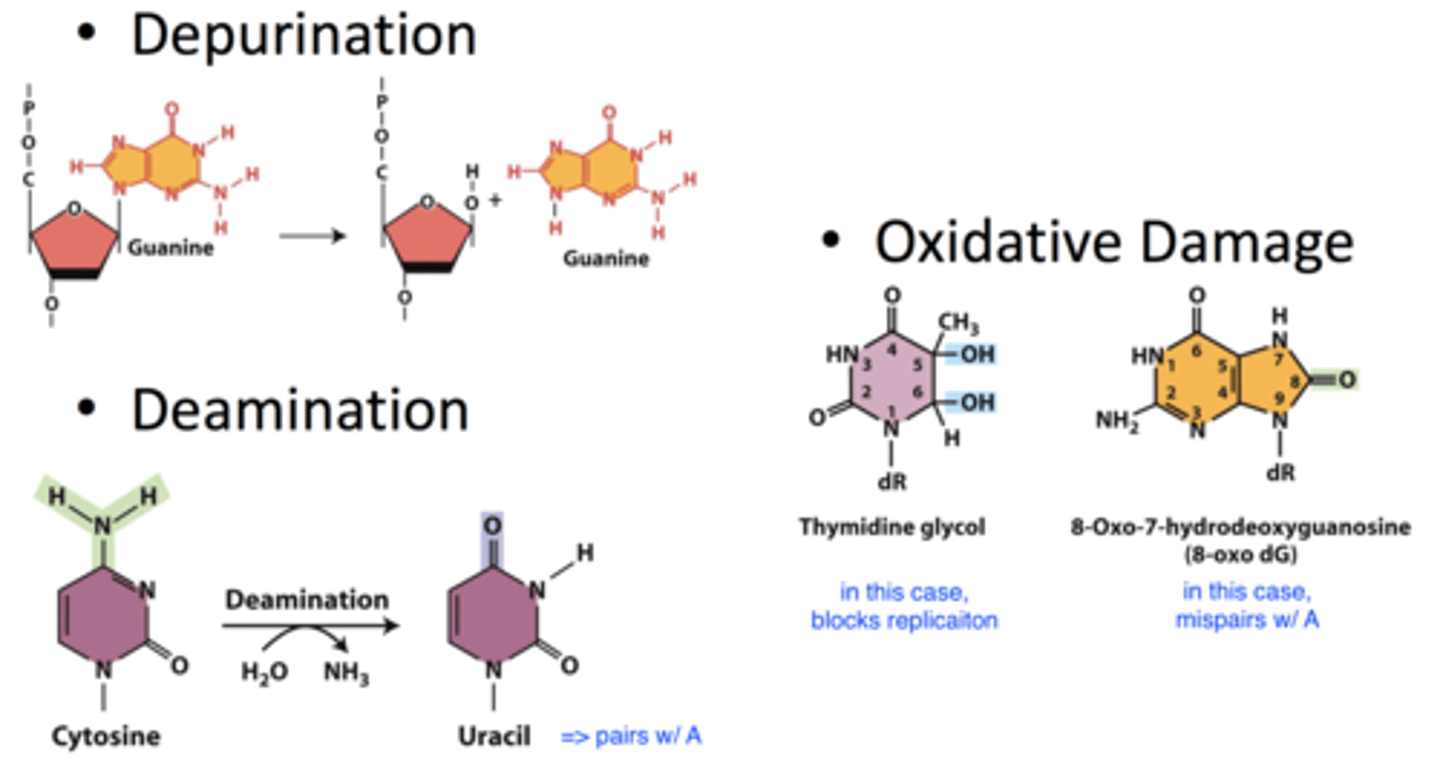
- depurination: lose purine base from DNA strand
-- occurs at a relatively frequent rate spontaneously
-- depyrimidation doesn't really happen
- deamination: remove amine group; converts cytosine to uracil which changes pairing
-- C-G => T-A mutation
- oxidative damage: occurs due to free radicals in cell
mutations can be induced by _______________
mutagenesis
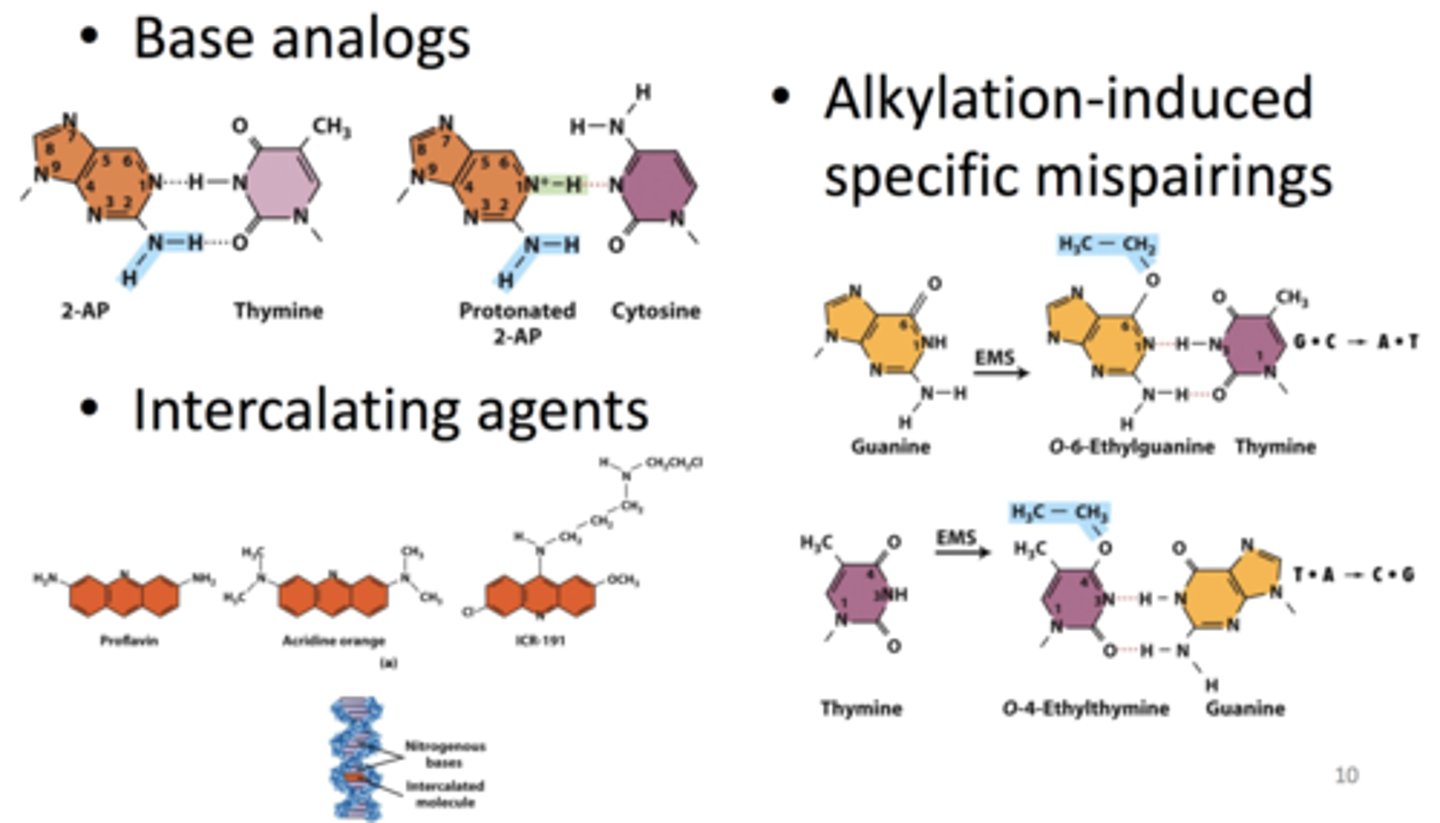
- base analogs: look similar to normal nucleotide bases but have slight differences
-- differences allow for different types of base pairing (ex. protonated vs non-protonated 2AP)
-- AT => GC or GC => AT transition
-- ex. 2-AP, 5-BU
- intercalating agents: ring structures model planar part of nucleotide bases; insert between stacked bases in a DNA molecule
-- cause insertion or deletion of nucleotides (frameshift)
-- ex. proflavin, acridine orange, ICR-191
- alkylation-induced specific mispairings: alkylation of nucleotide base prevents base from base pairing normally
-- O-6-ethylguanine pairs with thymine (GC => AT transition)
-- O-4-ethylthymine pairs with guanine (TA => CG transition)
- hydroxylamine: base modifier
-- GC => AT transition
base damage often causes _______________
a replication block
- UV induced base damage
-- can cause cyclobutane pyrimidine dimer or 6-4 photoproduct
- X-Ray/Ionizing radiation: backbone breaks and/or base damage
-- double strand breaks = chromosomal rearrangements
-- base damage = apurininc or apyrimidinic site (damage releases base portion of site)
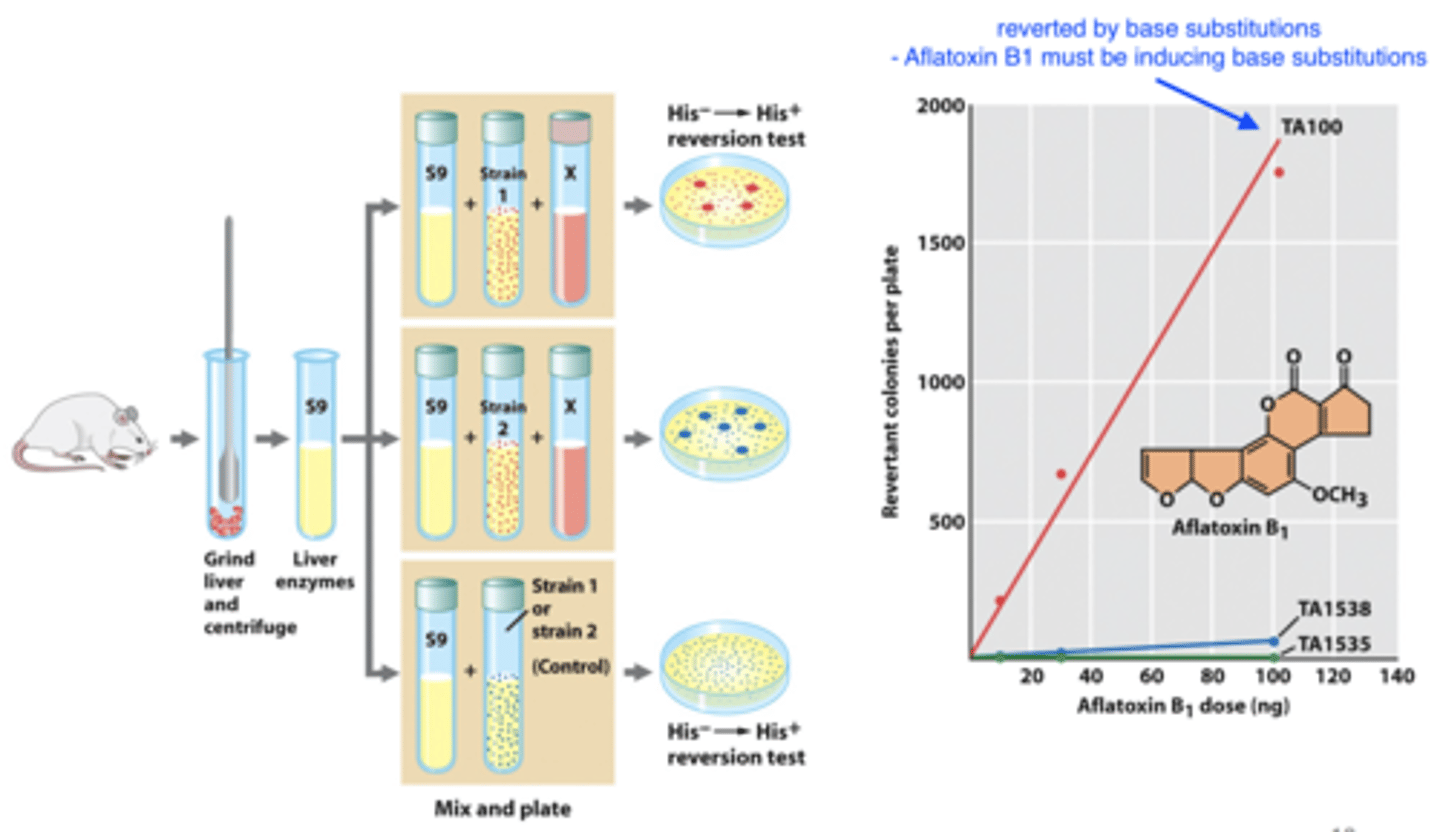
determining whether or not a substance is mutagenic
- examine ability to cause mutations (Ames' test)
-- first level of detection of potential carcinogens
- assess compound and its metabolites by including liver enzymes
-- tells us whether metabolites are mutagenic
- determine type of mutation induced
-- use his strains of salmonella typhimurium
--- TA100: reverted by base-substitutions
--- TA1538 & TA1535: reverted by indel mutations (insertion or deletion)
-- quantify number of his+ revertant colonies for each strain
Ames' test
1. isolate liver enzymes
2. grind up enzymes and add in bacterial strains and compound X
-- liver enzymes + bacterial strains = control (low % revertants)
-- if X is mutagenic we see revertants
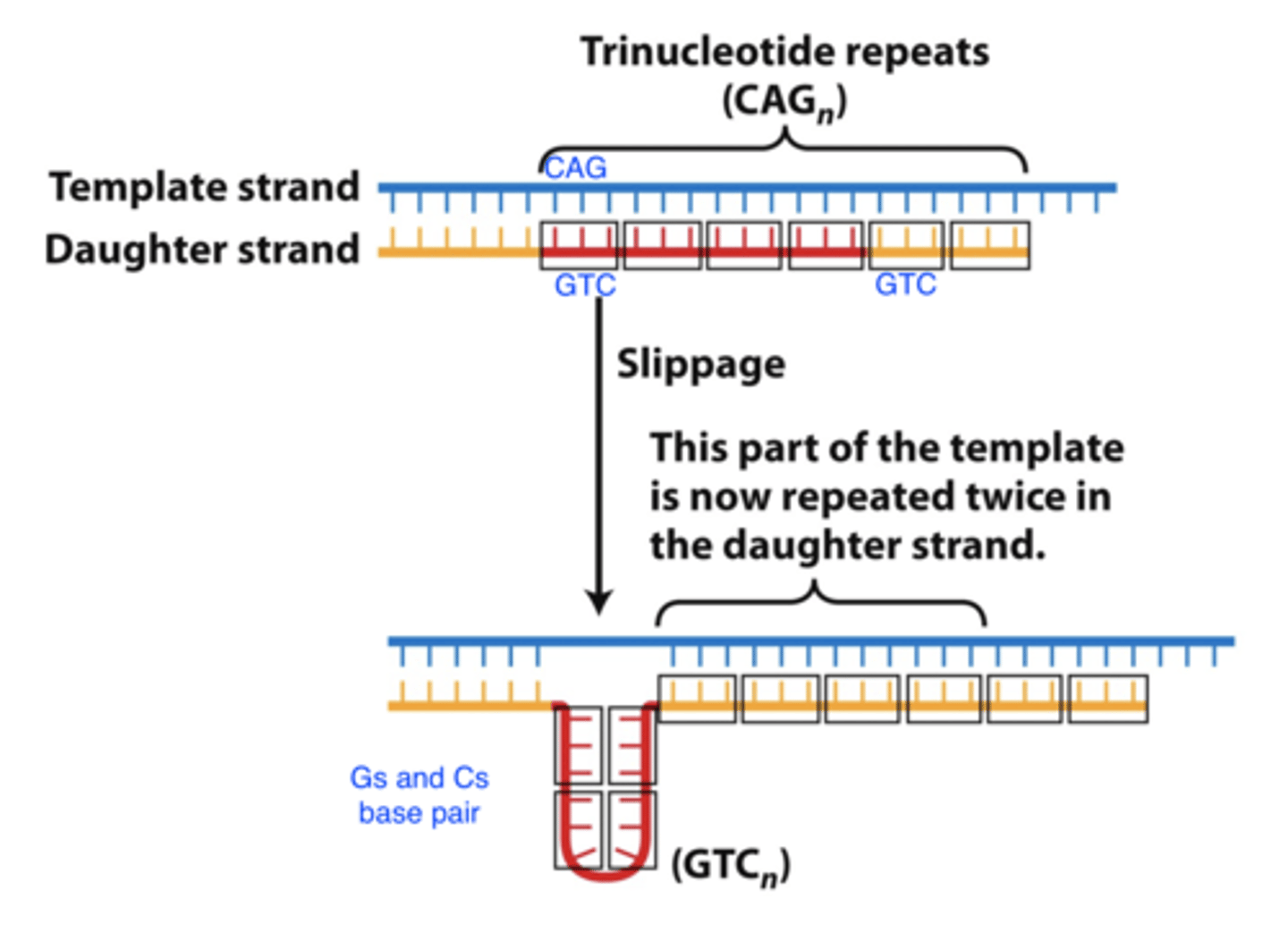
replication slippage
- replication slippage at repetitive sequences causes repeat expansions
-- in this example, leads to insertion of 4 GTC trinucleotides during replication
Huntington's disease
- autosomal dominant neurodegenerative disease
- molecular basis is expansion of a trinucleotide repeat
-- (CAG)n => Gln
-- poly Gln disease
-- normal: 10-35 repeats
-- disease: >36 repeats
fragile X syndrome
- CGG trinucleotide repeat located in 5' UTR of FMR1 gene
-- normal: 6-59 repeats
-- largely normal but prone to expansion from gen to gen (protein still produced): 60-200 repeats - can have fragile X associated tremor/ataxia syndrome (FXTAS)
-- affected: >200 repeats
- mutation in 5' UTR leads to:
-- methylation => prevents transcription => no FMRP protein production
-- loss of FMRP protein leads to fragile X phenotype
- mental impairment, seizures
- located on X chromosome
-- ~1 in 1,500 males
-- ~1 in 2,500 females
- can cause breakage of chromosome
damage in DNA sequences leads to...
repair of damage by DNA repair pathways (usually)
- DNA sequences are relatively stable despite frequent damage
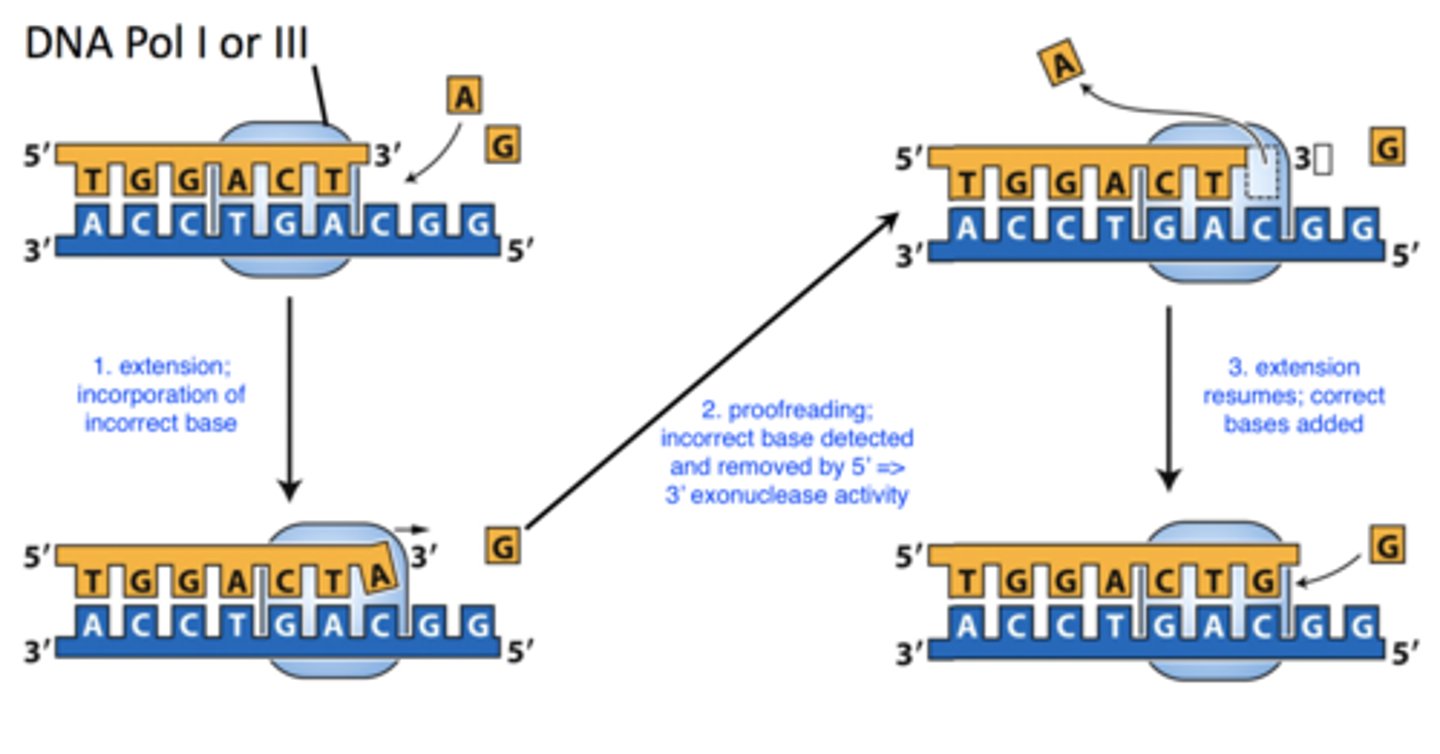
proofreading
- removes mispaired bases during replication
- DNA Pol I or III
-- both have 5' => 3' exonuclease activity
direct reversal of damaged DNA
- damage: mutagenic photodimer (cyclobutane pyrimidine dimer [CPD]) caused by UV light
- recognizes: base structure
- repair mechanism:
-- CPD photolyase: binds the photodimer & splits it to
regenerate the 2 original bases
-- photoreactivation: (enzyme requires light to function)
- BUT only repairs one very specific type of damage
![<p>- <b>damage:</b> mutagenic photodimer (cyclobutane pyrimidine dimer [CPD]) caused by UV light<br>- <b>recognizes:</b> base structure<br>- <b>repair mechanism:</b><br>-- <b>CPD photolyase:</b> binds the photodimer & splits it to<br>regenerate the 2 original bases<br>-- <b>photoreactivation:</b> (enzyme requires light to function)<br>- BUT only repairs one very specific type of damage</p>](https://knowt-user-attachments.s3.amazonaws.com/0c7b4888-d53c-4e7a-b75c-1585fecb91fa.png)
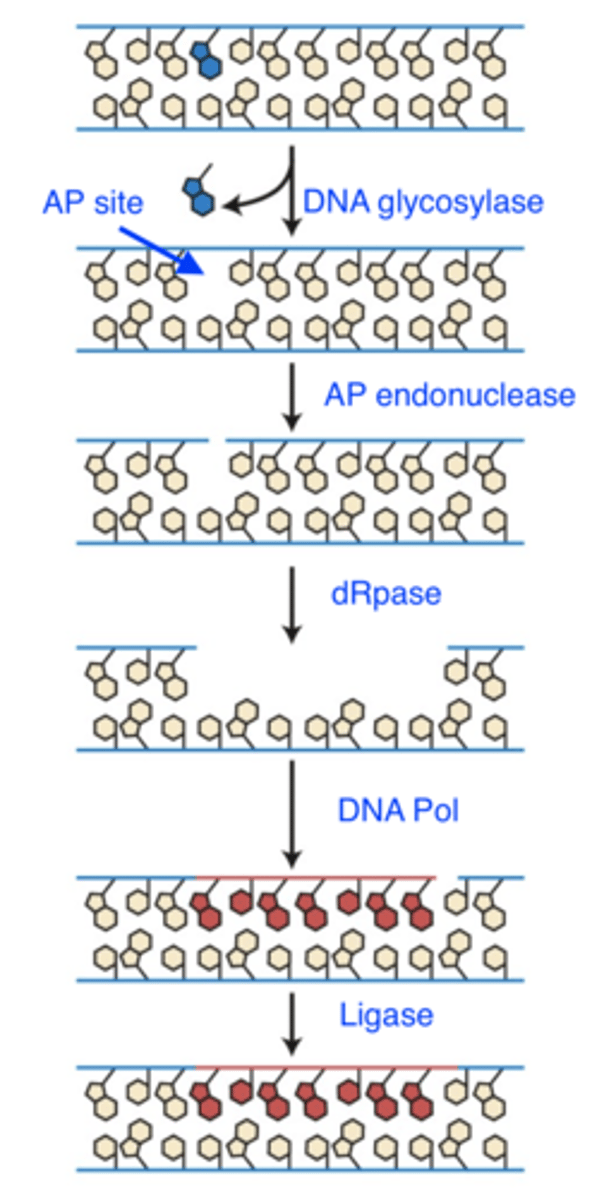
base excision repair
- damage: non bulky base damage
- recognizes: altered base structure
- repair mechanism:
1. DNA glycosylase cleaves base sugar bond
-- leaves AP site
-- can also fix existing APs
2. AP endonuclease makes cut in backbone
3. dRpase removes stretch of DNA
4. DNA Pol synthesizes new DNA
5. Ligase seals nick in backbone
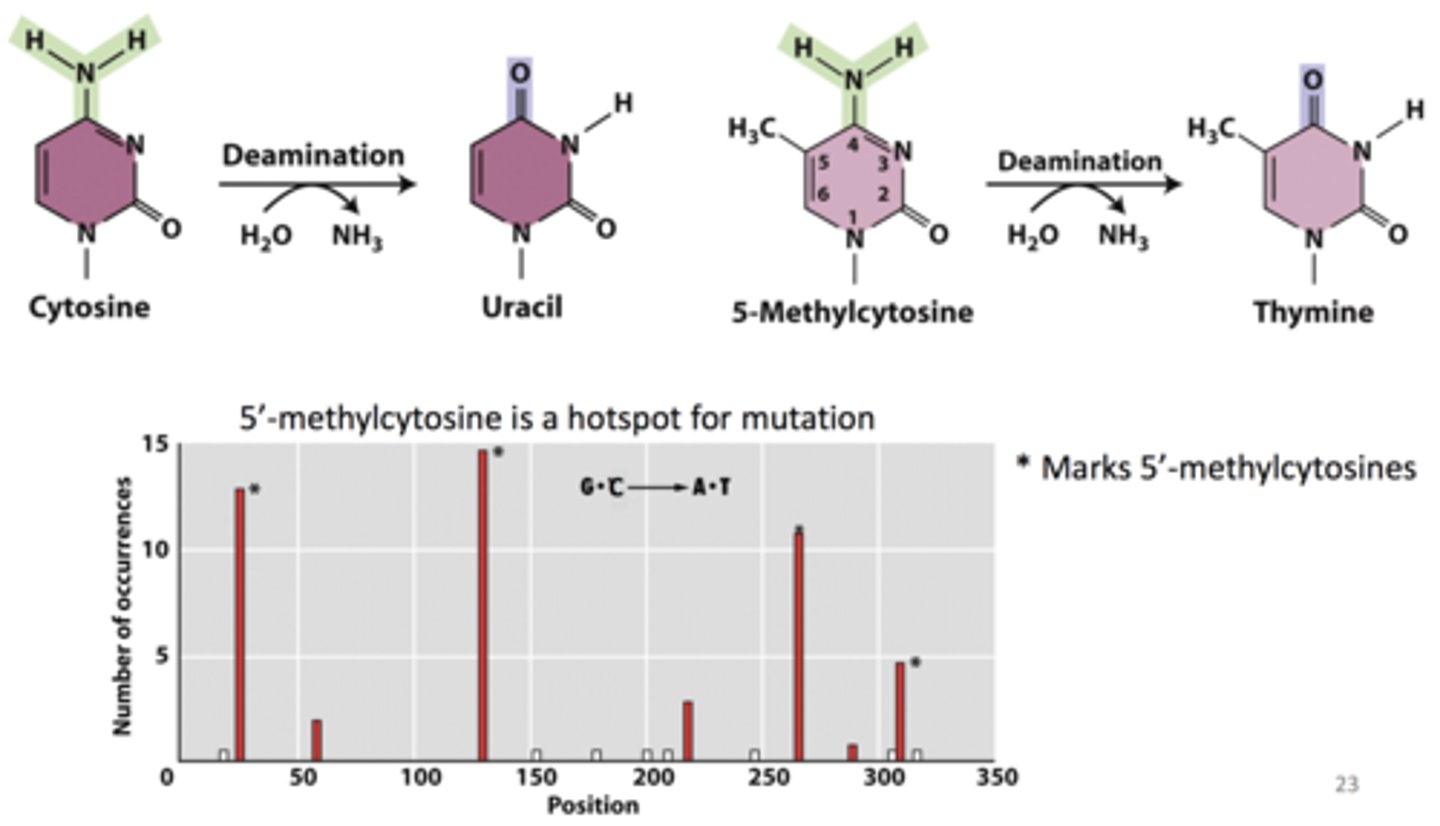
Uracil-DNA glycosylase
- removes uracil from DNA
-- deamination of cytosine => uracil (CG => UA)
- BUT deamination of 5-methylcytosine => thymine (CG => TA transition)
-- so... 5' methylcytosine is a hotspot for mutation
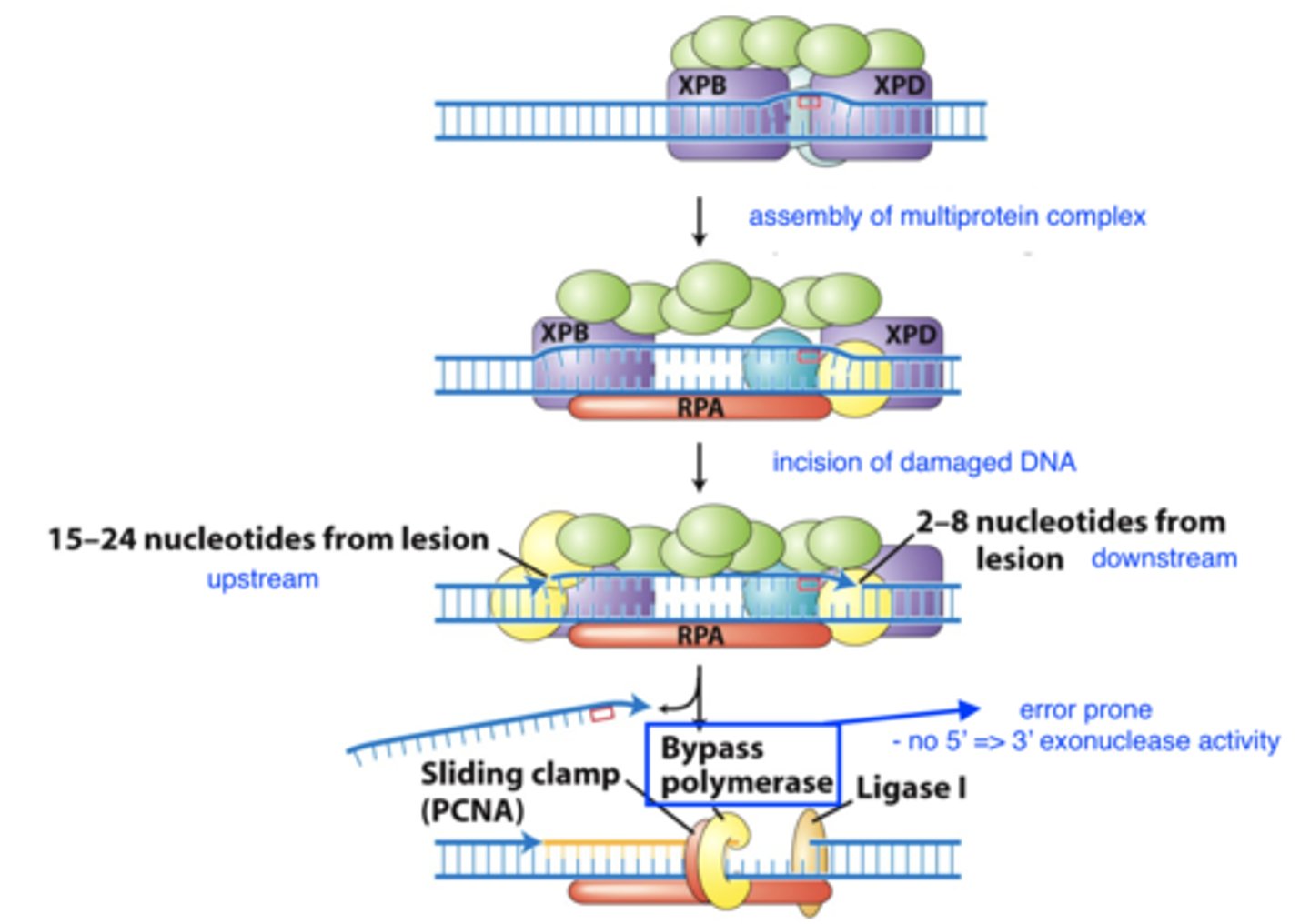
nucleotide excision repair
- damage:
-- bulky base damage or multiple damaged bases (replication stall)
-- abnormal or damaged base (transcriptional block)
- recognizes: stalled replication fork or transcription complex
-- uses base structure to identify damaged base
- general repair mech:
1. recognize damaged base
2. assemble multiprotein complex
3. cut damaged strand and remove nucleotides
4. DNA Pol copies undamaged template strand, followed by ligation
nucleotide excision repair recognition steps
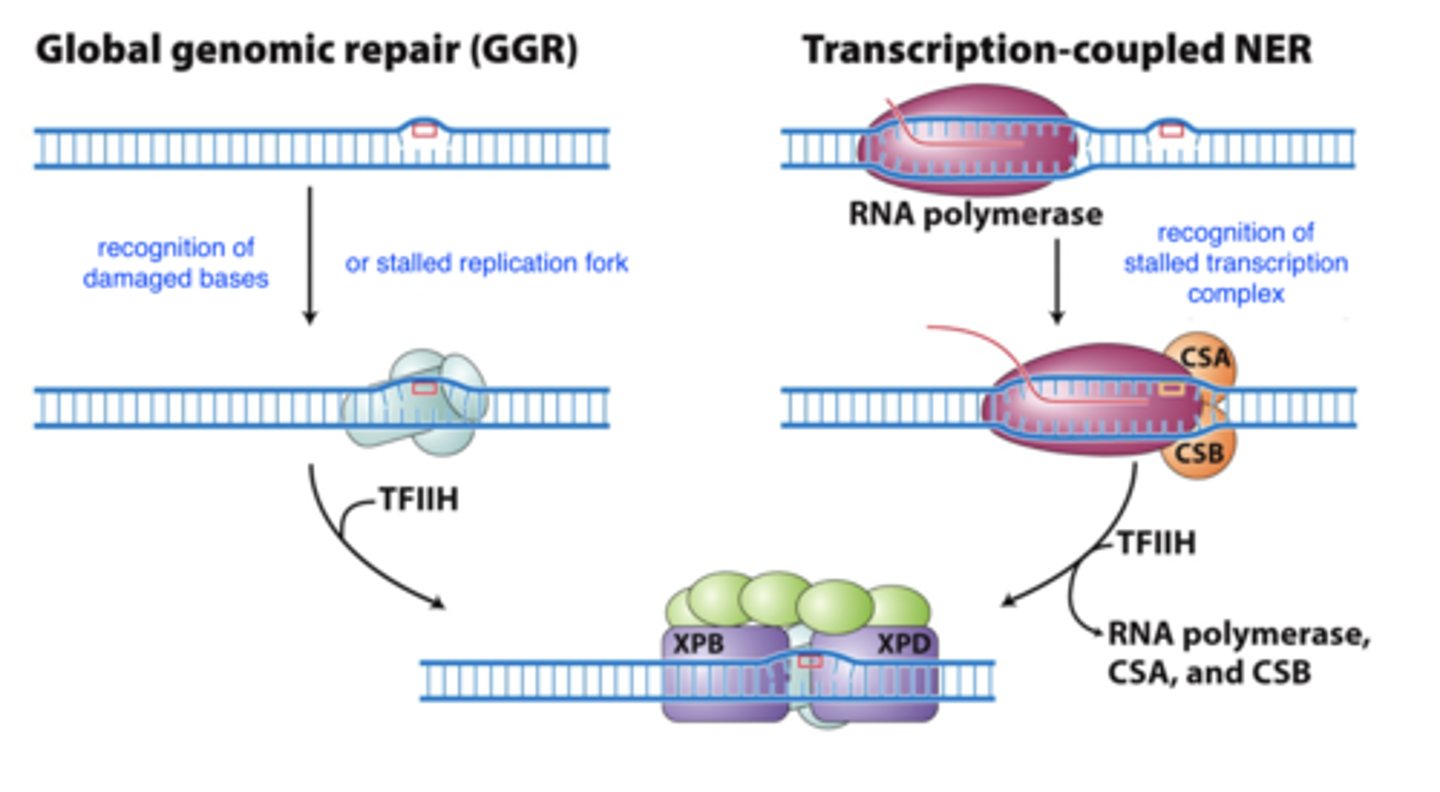
nucleotide-excision repair - repair mech
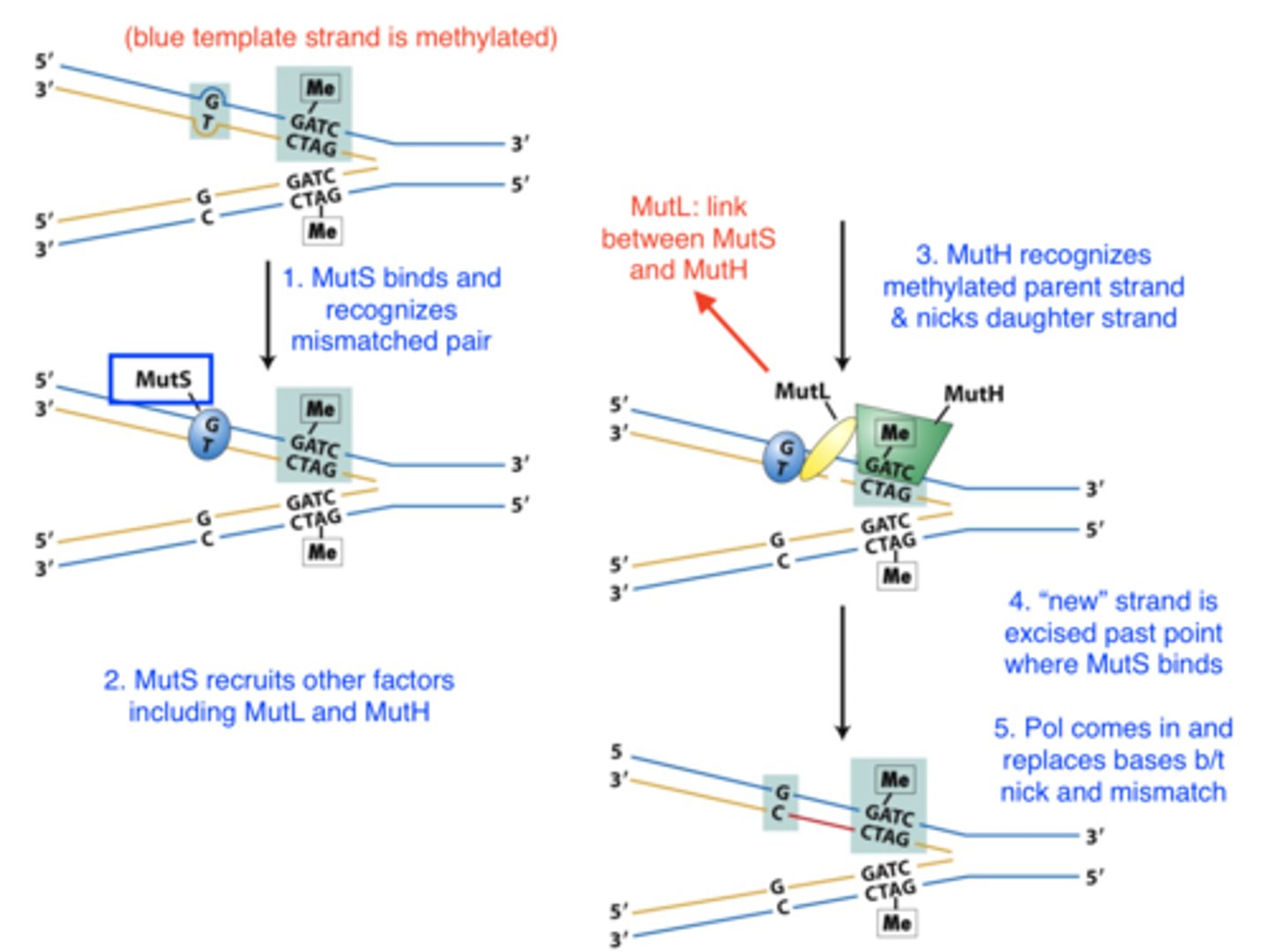
postreplication repair: mismatch repair
- damage: remaining replicative errors
- recognizes: mispaired bases
-- use DNA methylation to identify template strand (DNA becomes methylated after replication)
- repair mechanism:
1. recognize mismatched base pairs
2. determine which base in the mismatch is the incorrect one
3. excise the incorrect base and carry out repair synthesis
mismatch repair in E. coli
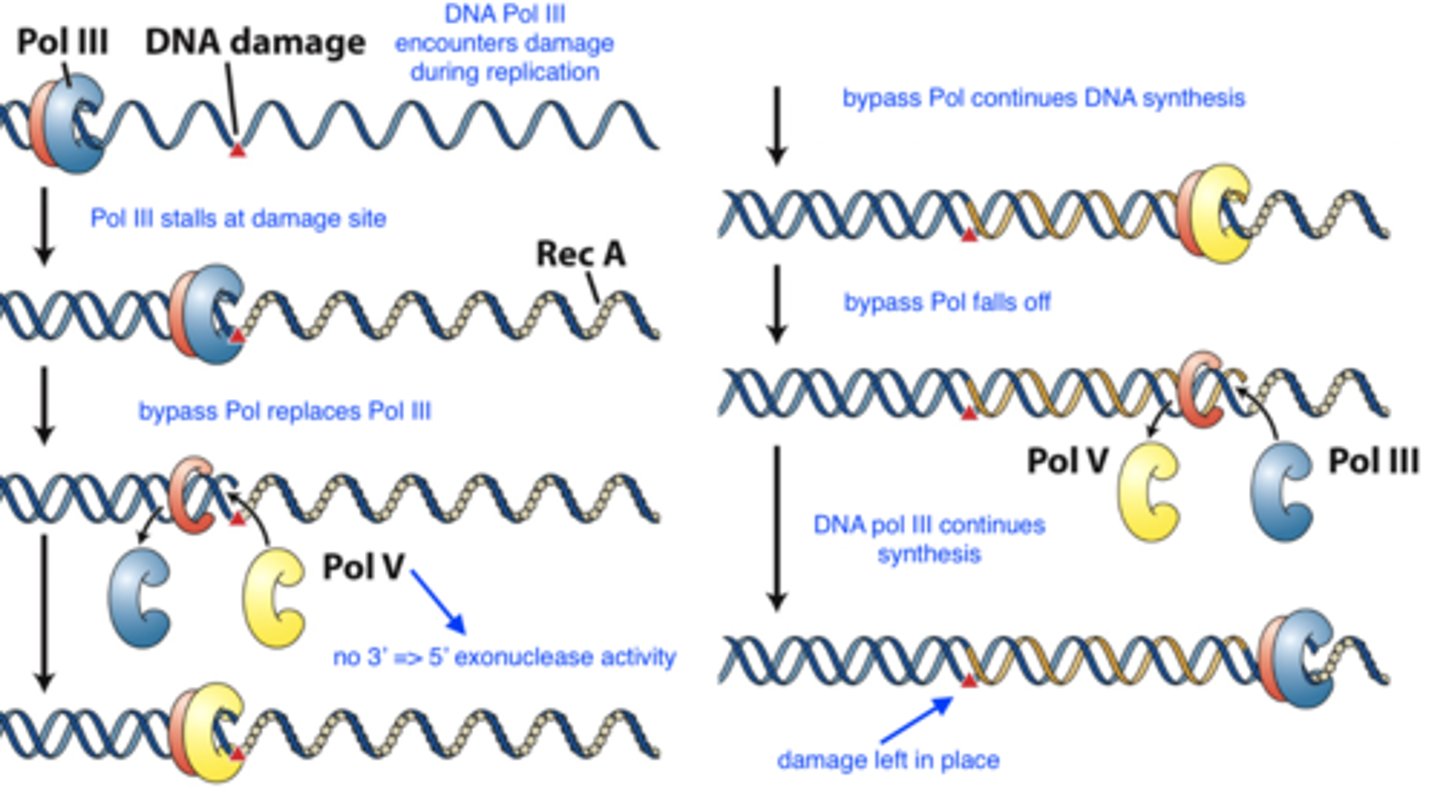
SOS system (translesion synthesis)
error prone
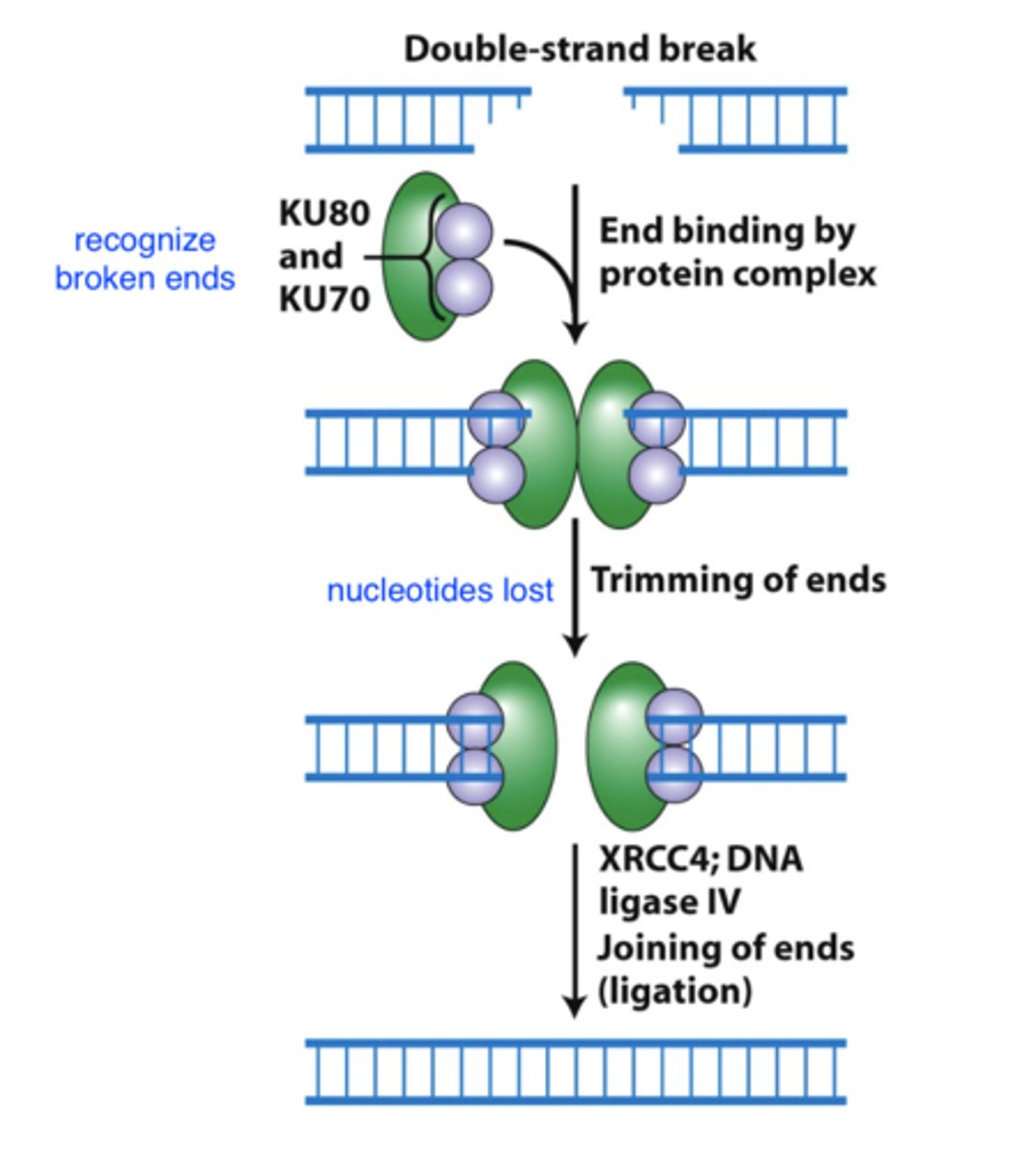
repair of double strand breaks by non-homologous end joining (NHEJ)
error prone
can lead to:
- deletions
- translocations
- inversions
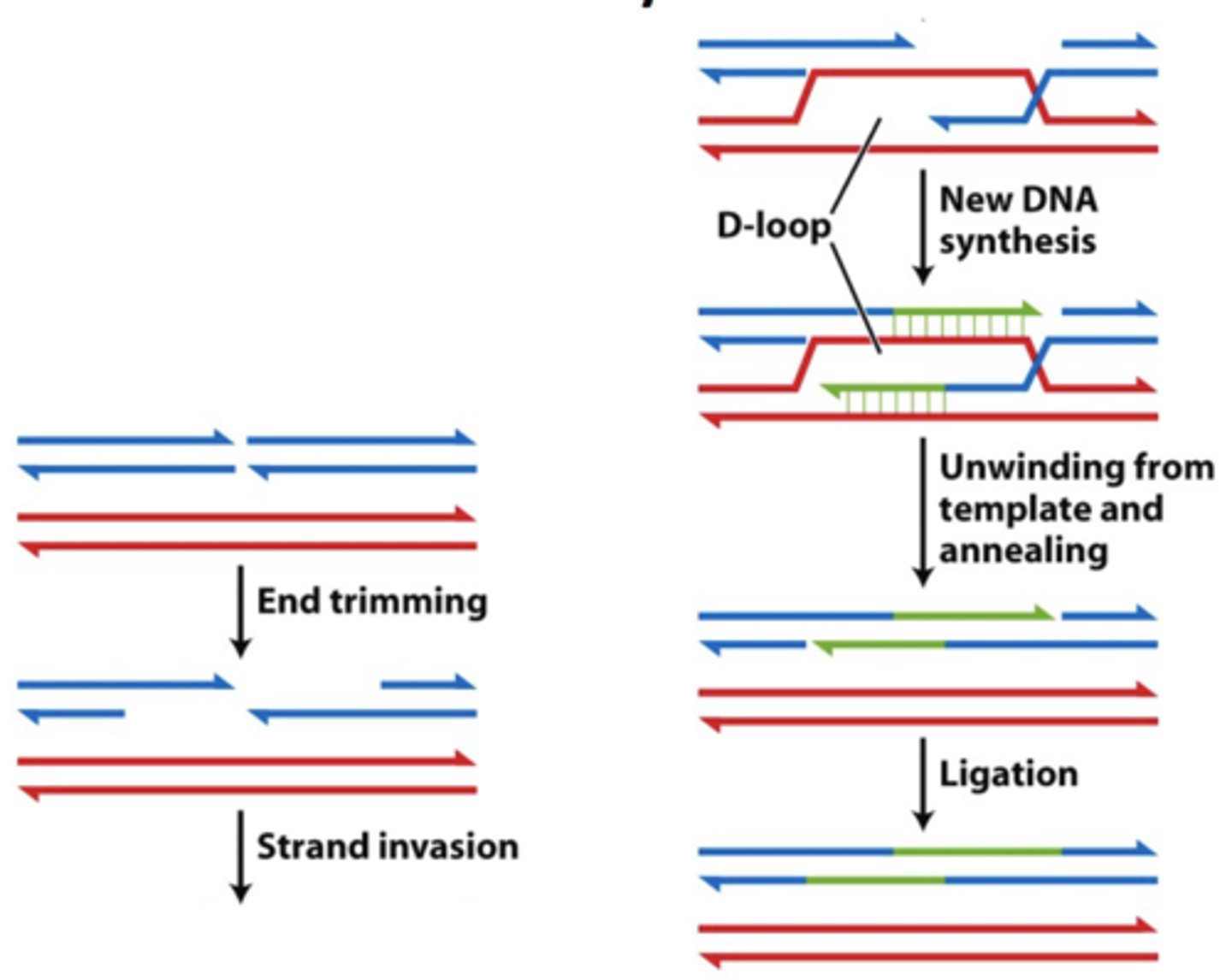
repair of double strand breaks by SDSA (synthesis dependent strand annealing)
"error-free"
- cell uses homologous chromosomes in order to repair break (relatively error free but can alter alleles)
-- homologous recombination
-- can also use sister chromatids (completely error free)
cancer cells
cancer: aggregate of cells, all descended from an initial aberrant founder cell
- characteristics:
-- rapid division/proliferation rate (cancer cells don't stop proliferation when in contact with one another like normal cells)
-- ability to invade new cellular territories
-- high metabolic rate
-- abnormal shape
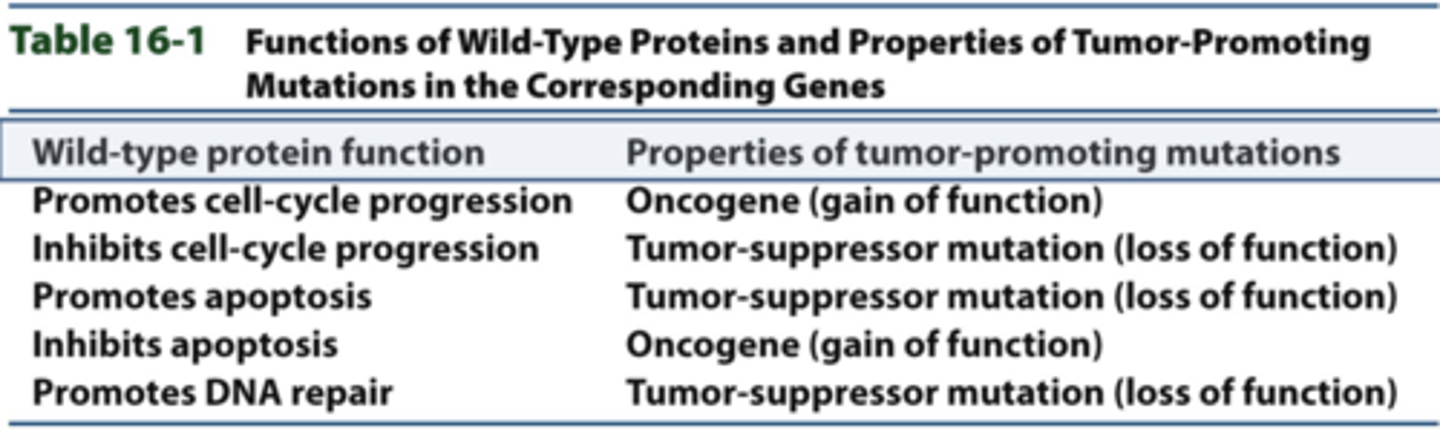
mutations in cancer cells
oncogenes:
-- gain of function
-- dominant
-- proteins encoded by oncogenes are usually activated in tumor cells
-- mutation need be present in only one allele to contribute to tumor formation
-- wt = proto-oncogene
tumor-suppressor genes:
-- loss-of-function (LOF)
-- recessive
-- encoded gene products lose much or all of their activity in tumor cells
-- mutation must be present in both alleles for cancer to develop
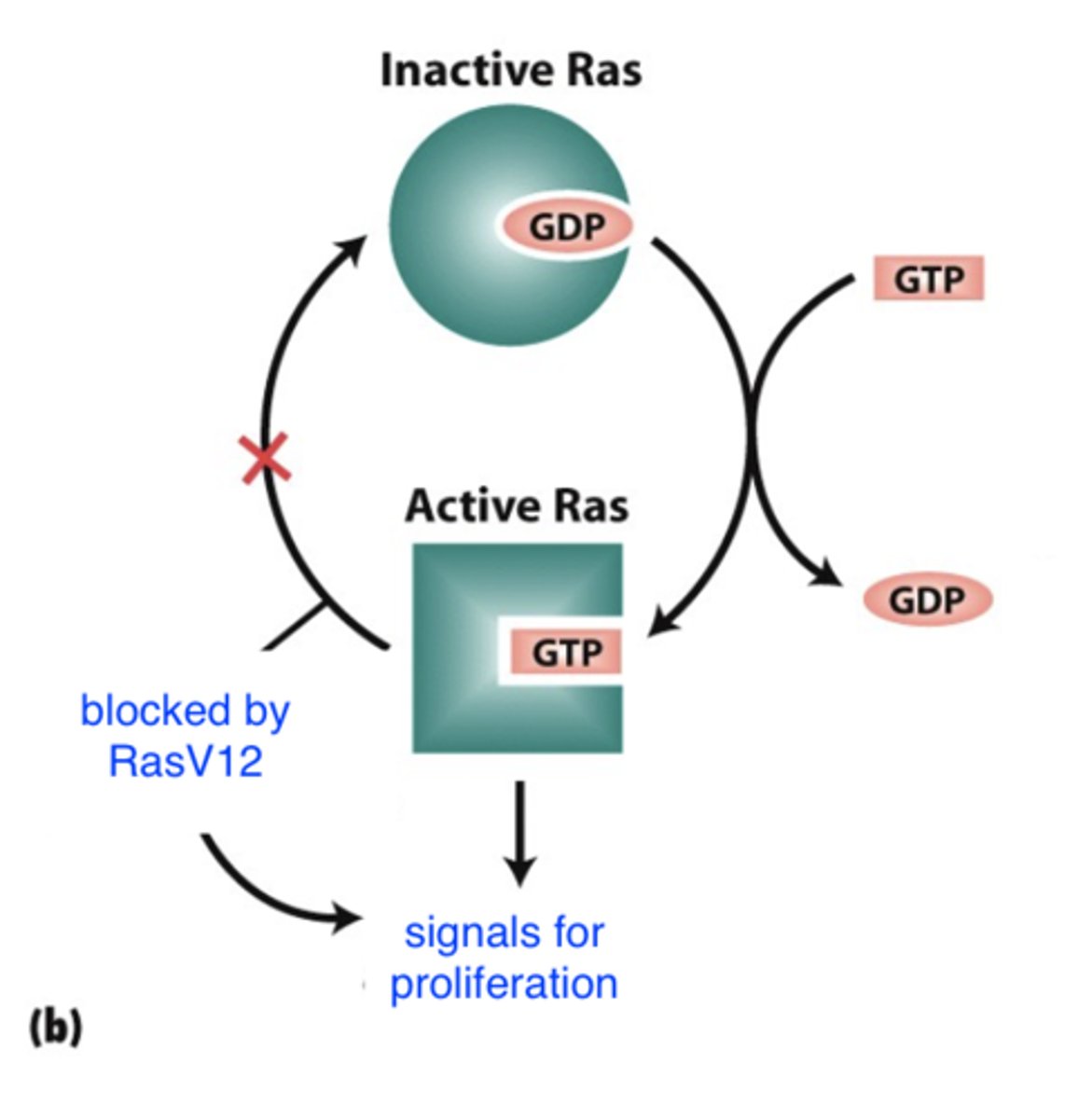
ras oncogene
- GTPase: active when bound to GTP
- continuously active
- proto-oncogene => oncogene via G => T transversion
-- Gly => Val
- normally ras has an active form and an inactive form
-- mutation blocks conversion of active Ras to inactive Ras
=> always signaling for proliferation
properties of tumor-promoting mutations
p53
- prevents progression of cell cycle until DNA damage repaired
- can induce apoptosis (cell death
=> tumor suppressor mutation
___________________ in ____________ chromosomes lead to cancer
rearrangements/translocations in somatic chromosomes
- relocation of an oncogene
- formation of a hybrid oncogene
- mutations in BRCA1 and BRCA2
relocation of an oncogene
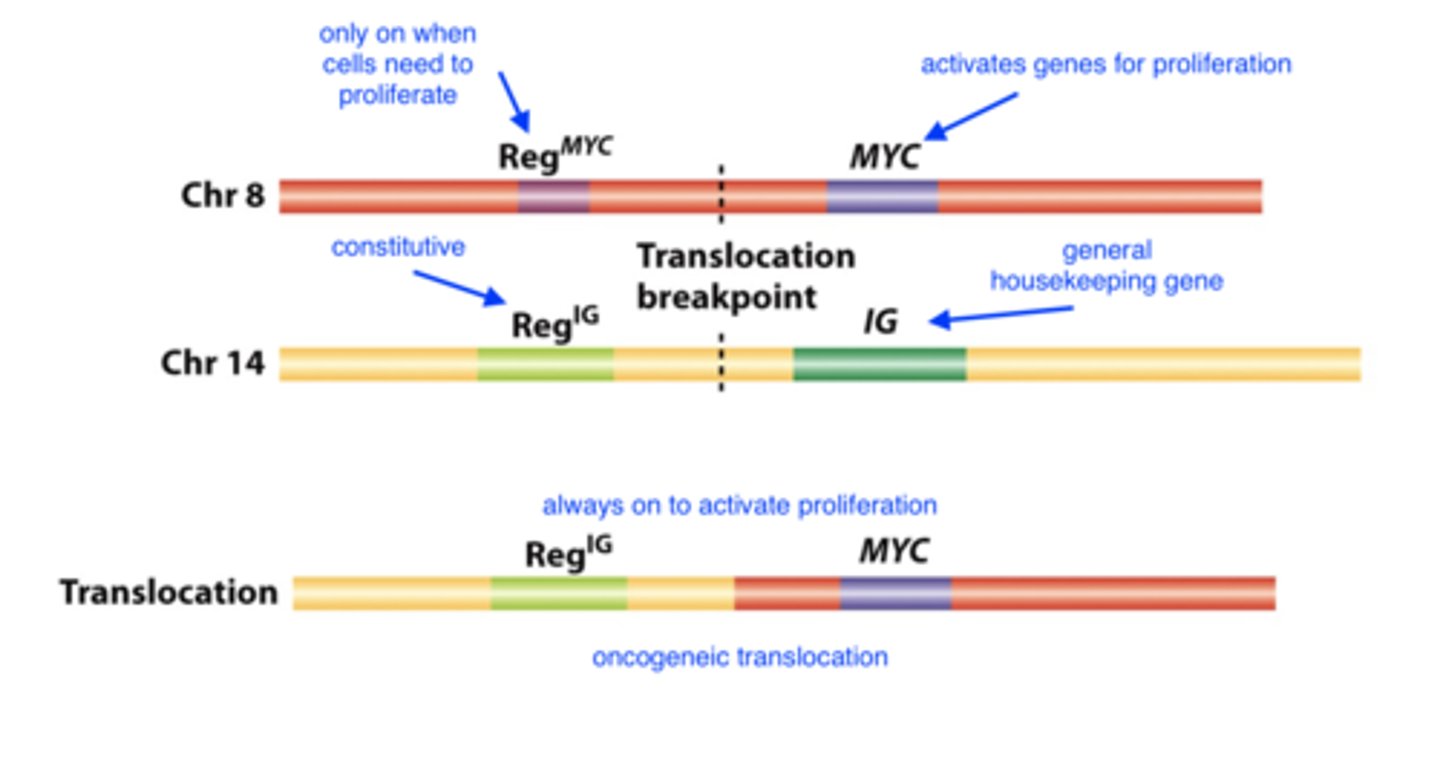
formation of a hybrid oncogene
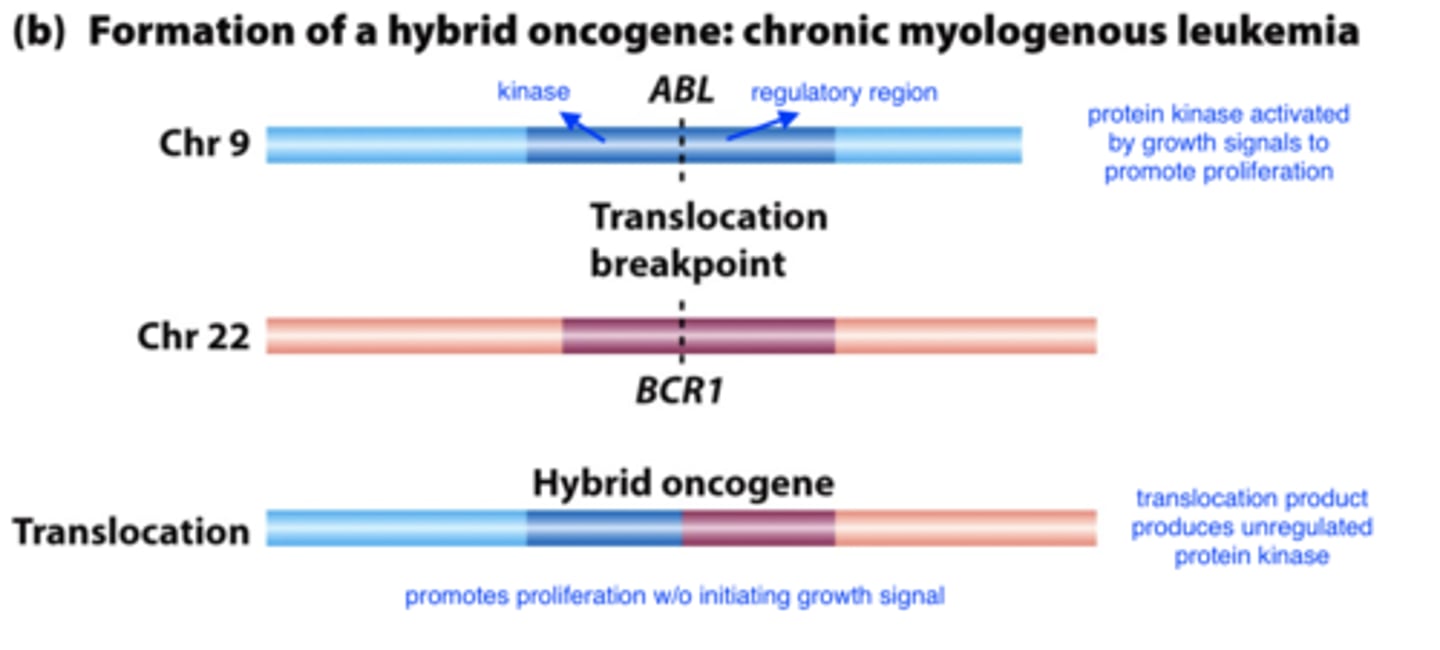
BRCA1 and BRCA2
- mutations lead to defects in repair by homologous recombination (and possibly other pathways)
-- both BRCA1 and 2 bind Rad51 (REcA homolog)
-- Rad51 catalyzes DNA strand invasion during HR
-- BRCA1/2 act as a scaffold
- heterozygous mutations in these genes greatly increase the chance of breast/ovarian cancer in patients