Lecture 5 - Prokaryotic Transcription
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
Mechanism of Transcription
coupled transcription and translation (happen simultaneously)
once the 5’ end of the RNA is accessible, the ribosome binds to initiate translation
requires a DNA template, 1 RNA pol, and NTPs
The progress of the ____ and the ____ are linked in prokaryotic transcription.
polymerase; ribosome this is because transcription and translation are coupled
Steps of Transcription
initiation
elongation and proofreading
termination
The Promoter
collection of DNA sequence elements, including core promoter and promoter proximal elements, that are required for initiation of transcription OR that increase the frequency of initiation only when positioned near the transcriptional start site
The elements of DNA are defined in relation to the ____ ____ ___, which is denoted as +1.
transcription start site (TSS)
we refer to the features in the sequence as being upstream or downstream (to the right is upstream)
Bacterial Promoter Sequences
not absolutely conserved but do show consensus sequence
the strength of the promoter (how efficiently and the frequency with which it initiates transcription) is proportional to how close the bacterial promoter is to the consensus
basically a strong promoter is the one that is the closest to the ‘ideal’ sequence since it binds RNA polymerase better
A strong promoter have a better ____ for the RNA polymerase.
affinity;
consensus sequences in the promoter include:
-10 sequence TATA box → TATAAT
-35 sequence TTGACA
the spacing between the -10 and -35 sequences, and the TSS is important → deletions or insertions that change the spacing are deleterious some bacterial genes have UP element (found in RNA and essential genes)
Some bacterial genes have ____ element.
UP; found in RNA and essential genes
Factors that Aid Transcription Initiation
point is to make DNA strand separation easier:
AT rich sequences (like at the -10 sequence) are easier to melt because of the differences in base stacking and H-bond interactions (compared to GC rich sequences)
negative supercoiling loosens alpha-helix easier strand separation
RNA Polymerase
can synthesize RNA de novo (all by itself, DNA can’t do this)
bacterial polymerase is a holoenzyme considered comprised of the core RNA polymerase and a sigma factor (sigma54 RpoN or sigma70 RpoD)
core enzyme is made of 5 subunits and weights 400kDa
Sigma Factor
part of the bacterial polymerase holoenzyme
can be one of two protein families: sigma 54 RpoN and sigma70 RpoD
sigma factor is responsible for:
directing the holoenzyme to specific promoters
melting the -10 region, and stabilizing
interact with other transcription
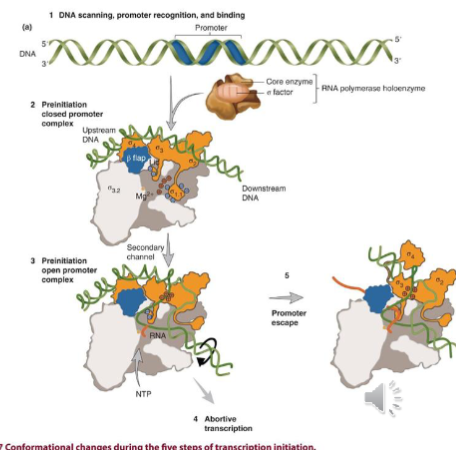
Steps of Transcription Initiaiton
RNA pol spends 85% of its time searching for promoters - it binds once it finds a promoter
holoenzyme binds the promoter at -35 and -10 to form the closed preinitiation promoter complex. this step is reversible and the DNA is double stranded (makes a closed structure)
complex undergoes a structural transition to the open preinitiation promoter complex - wherein approximately 12-14 base pairs around the transcription start site (-10 area) are melted to expose the template strand of the DNA and form the transcription bubble
usually irreversible and involves the presence of NTPs (no NTPs around = nothing happens)
abortive transcription: characterized by production of short transcripts (2-15 nucleotides) in length and a long pause where the pause is regulated (revving before greenlight)
promoter escape: aka promoter clearance; current model has portions of the sigma factor being displaced to allow for exposure of the RNA exit channel
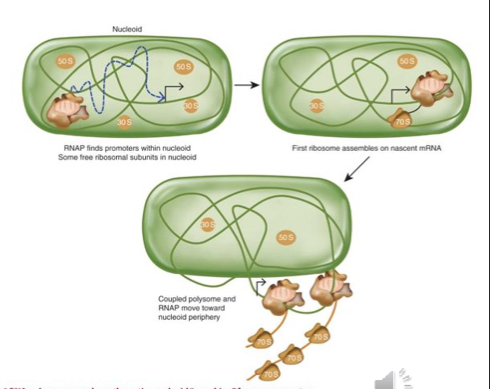
Elongation
starts after about 8 nucleotides are synthesized (is temporarily an RNA:DNA hybrid at this point)
transcription continues 5’-3’ - most essential and highly transcribed genes are oriented in the same direction as replichore (origin and terminus of replication) to decrease torsional stress
one replisome goes clockwise, other goes opposite way
RNA pol makes a footprint of an area of around 30b protected from nucleases
Elongation and Proof-Reading
as polymerase elongates, there is an RNA:DNA hybrid of 9-12 bp in the transcription bubble - starts and pauses, which allow for the ribosome to catch up
the high fidelity is due to the proof-reading activity
if the RNAP incorporates the wrong incorrect NTP:
it backtracks on the DNA template, this motion moves the nascent RNA out of the active site
after backtracking for correction ~5 nucleotides the pol pauses and recently added bases are cleaved off by nuclease activity while in the backtracked state
pol resumes polymerization with new 3’ end available from the cleavage event
e. coli RNA polymerase has high fidelity and proofreading activity
RNA pol adopts conformations related to its function based on the stage of transcription (T and R)
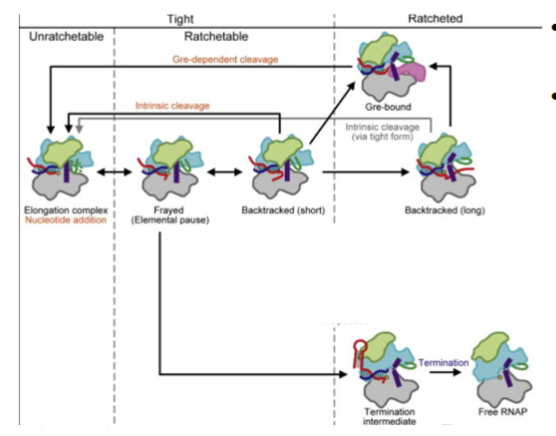
Tight Form RNAP
happens in elongation for GreA-dependent RNA cleavage, arrest, and hairpin-pause/termination
unratchetable nucleotide addition
ratchetable intrinsic RNA cleavage
Ratcheted Form RNAP
happens in initiation
gre-dependent RNA cleavage nucleotide addition, and its reversal e.g. during abortive transcription
Transcription Termination
can be Rho dependent or Rho independent
Rho independent termination is considered intrinsic
Rho dependent termination is reliant on presence of Rho to stop transcription - without it you get read-through (continues to read past where gene should’ve ended)
most bacteria have rho gene and it is essential in E.coli but there are some that don’t have it
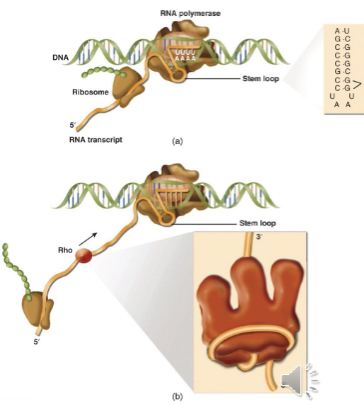
Rho Independent Termination
consensus inverted repeat makes a hairpin/stem loop structure that destabilizes the transcription bubble; termination is structure and sequence based
forms in the mRNA just before the last nucleotide is added and destabilizes the transcription bubble
is then followed by transcription of a string of 7-7 uracils in the RNA (remember that nascently transcribed RNA in the bubble is base paired with the DNA and this interaction has the lowest stability of the complementary base pairing)
combination of these two leads to polymerase pause and transcript release
Consensus Sequence
aka canonical sequence, the ‘ideal’ form of a DNA sequence that is found in slightly different forms in different organisms but believed to have the same function
gives the nucleotide most often found for each position
What does Rho do?
mediates release of the transcript at a DNA sequence at which the transcription complex is too stable for spontaneous release
Catch Up Model
prevailing model for Rho-dependent transcription termination
Rho binds specifically to an 80-90 nucleotide long C-rich sequence (Rho utilization, or rut site) as the newly formed RNA emerges from the exit site of RNA pol
Rho temporarily releases one of its subunits to then have RNA placed in the middle of the ring
Rho travels 5’-3’ chasing the polymerase in an ATP-dependent process. When the
polymerase stalls at the terminator stem‐loop structure, Rho catches up and “traps” the polymerase by inducing a shape change that inactivates the catalytic center of the polymerase.Rho then unwinds the weak DNA–RNA hybrid. This unwinding causes termination of RNA synthesis and release of all the components.
The tsp region can extend upto 100b to give room for the Rho to “catch up and trap” the polymerase
There are factors that aid to uncouple transcription and translation to allow for the Rho to gain access to the RNA
Rho Dependent Termination
no consensus sequence but there IS also an inverted repeat except it doesn’t have a run of Ts in the non-template (no run of As in the template)
termination facilitated by the Rho protein (hexameric ring protein that has RNA helicase and RNA translocase activity)
translocase activity helps move the protein along by either pulling RNA through or pushing the protein forward
Rho-dependent terminator consists of two parts, rut region and tsp region (where transcripts are terminated)
tsp Region
transcription stop point; the downstream sequence in a Rho-dependent terminator where transcription is physically terminated
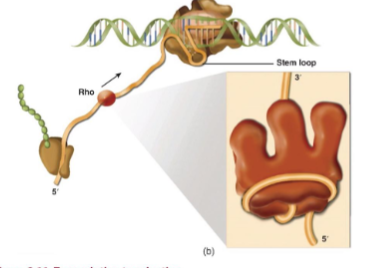
Stand By Model
Rho binds with RNAP aided by a NusA (transcription elongation factor) in noncoding or untranslated regions of nascent RNA, making a pre-termination complex (PTC)
Rho scans growing RNA chain for Rut site while in the PTC
it then triggers allosteric inactivation of the elongation complex and destabilization of the predetermination complex when Rut site is detected
once this happens Rho rearranges its open-ring conformation to capture it
Rho continues to widen the clamp associated with displacing the RNA 3’ end from the RNAP catalytic site
Contextualizing Transcription
transcription is part of gene expression but we have to control when and how quickly genes are transcribed → needs something to sense the change and activate the response to external changes
prokaryotic genes are generally arranged in operons
Operons
multigene transcriptional unit, including structural genes and control elements in DNA recognized by regulatory gene products
genes in an operon are transcribed from a single promoter to make a pre-mRNA (single primary transcript) or polycistronic RNA (many genes with multiple start and stop codons)
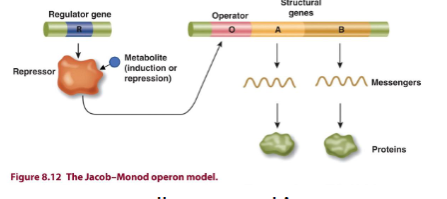
lac Operon
bacteria use glucose as a food source, but when glucose is scarce they have the ability to metabolize alternative sugars such as lactose
is a catabolic/inducible operon that responds to a change in environment
made of 3 structural genes:
lacZ (beta-galactosidase)
lacY (lactose permease)
lacA (transacetylase)
regulated:
LacI (lac repressor) - trans-acting factor
Pi (promoter for the lac repressor)
CAP site (for the catabolic activator)
Plac (promoter for the structural genes)
O (lac operator)
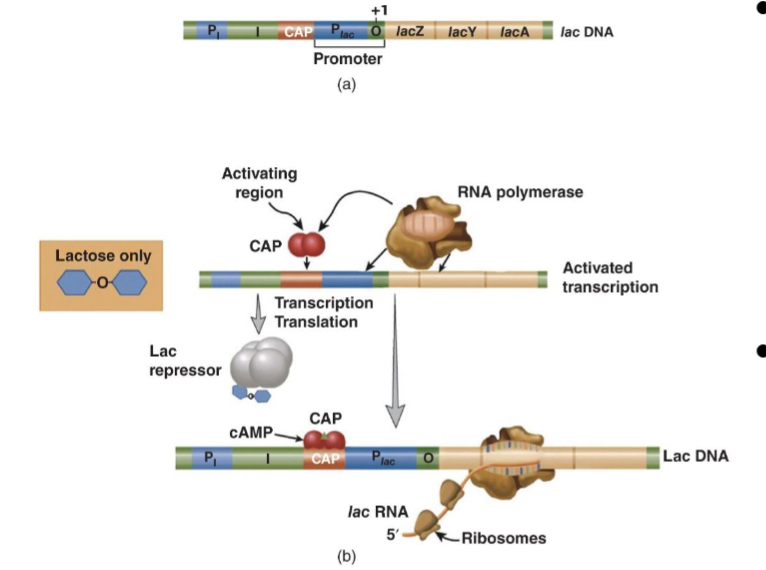
lac Operon Induction
lac repressor is constitutively transcribed (always expressed) and binds as a tetramer to lac operator O (+1 position) in the absence of lactose → blocks RNAP binding or initiation
generally repressing most of the products from the lac operon but there is a low level of basal transcription
lac repressor is a trans acting factor binding to the lac operator which is a cis acting sequence
trans acting factor: regulatory protein or factor that binds cis element to regulate gene expression
cis acting sequence: regulatory sequences of DNA/RNA that regulate gene expression by being bound by trans acting factor
turns on when glucose is unavailable and lactose is available - allolactose accts as an inducer
transcription is activated by CAP or cyclic adenosine monophosphate receptor protein which recruits RNA pol to the lac promoter: cooperative binding
The lac operon is induced when ____ is available and ____ is unavailable.
lactose; glucose
allolactose (made by transglycosylation of lactose by beta-galatosidase) is the inducer for the lab operon → when lac repressor is bound to allolactose it goes through a conformational change that decreases its DNA binding affinity thus relieving lac repression
transcription of the operator is activated by CAP or cyclic adenosine monophosphate receptor protein (CRP) which recruits the RNA polymerase to the promoter → cooperative binding
RNA pol then transcribes a polycistronic mRNA with start/stop codons for each of the 3 encoded proteins
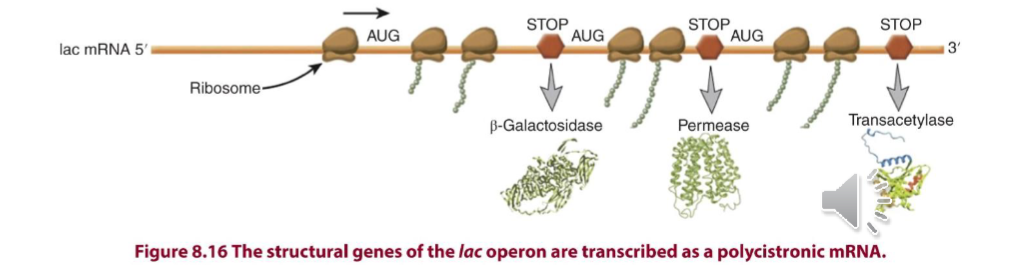
Cooperative Binding
process where the binding of a ligand to one site on a molecule affects the binding of subsequent ligands to other sites on the same molecule
happens in lac operon induction when CAP/CRP binding recruits RNAP to the lac promoter
Types of Operons
lac operon - catabolic/inducible operon
catabolic operons: expression of enzymes involved in catabolism of energy source
anabolic (biosynthetic operons): expression of enzymes in involved in synthesis of energy source
trp Operon of E. coli
tryptophan operon is negatively regulated by TrpR aporepressor
biosynthetic (anabolic) operon does de novo synthesis of tryptophan when needed
repressor protein is aporepressor (inactive when corepressor is gone)
5 structural genes code for enzymes responsible for synthesizing L-tryptophan
transcribed from a single promoter called Ptrp and a single strand of polycistronic RNA
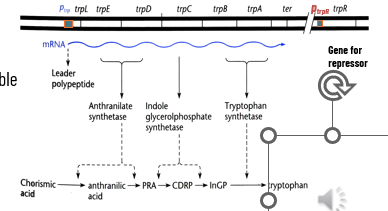
Negative Regulation of trp Operon by TrpR
negative regulation means default on and needs to be turned off
tryptophan needs to be made: structural genes are transcribed when promoter is available to RNAP - the aporepressor cannot bind to the operator site
when there is enough tryptophan, synthesis needs to be stopped - aporepressor and corepressor (trp) form functional repressor dimer and bind to the promoter to turn trasnscription off
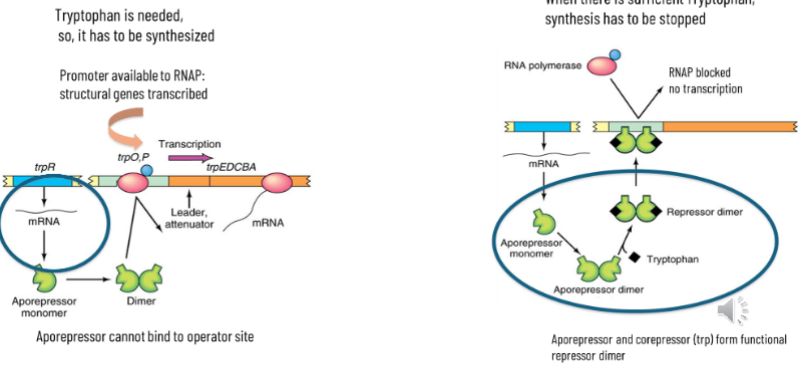
Autoregulation
tryptophan regulates its own production and regulates the action of the aporepressor
repressor dimer inhibits transcription of trp operon and transcription of the TrpR gene (the repressor gene)
2 Styles of Negative Regulation
lac operon: default state is an active repressor → active repressor and inducer (lactose)
needs active repressor and inducer for an inactive repressor (inactive repressor is allolactose)
trp operon: default state is on - tryptophan binds to TrpR and operator and turn off transcription
tryptophan is the effector and corepressor
inactive repressor is the aporepressor
aporepressor and corepressor makes the active repressor
High _____ of initiation of transcription describes a strong promoter.
frequency; a strong promoter has a high affinity for RNAP and a consequence to that is it’ll transcribe more
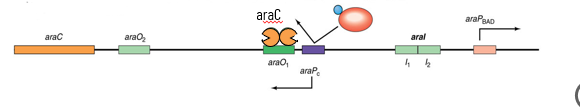
L-ara E. coli Operon
example of negative and positive regulation but was one of the first examples of positive regulation in bacteria
synthesis of L-arabinose (5C sugar), as a source of energy therefore catabolic
3 structural genes all transcribed from one promoter, Pbad:
araA
araB
araD
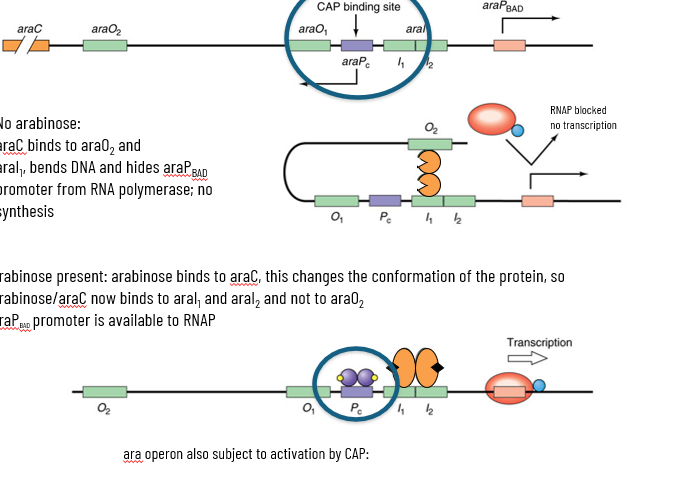
2 operators (araO1 and araO2) do repression
AraC regulator is the repressor that binds upstream one of the cis element upstream of the promoter
is also the activator upon binding of arabinose binds at a DIFFERENT set of cis elements upstream from promoter
operon is activated through cAMP/CAP/CRP
L-ara with and without Arabinose
no arabinose: araC binds to araO2 and araI1 bends DNA and hides araPbad promoter from RNAP; no synthesis
arabinose present: arabinose binds to araC, this changes the conformation of the protein, so arabinose/araC now binds to araI1 and arai2 and NOT araO2
araPbad promoter is available to RNAP
AraC Autoregulation
too much araC protein → binds to araO1 operator to prevent further transcription (RNAP cannot bind to araPc)
allosteric change in regulatory molecules as a result of small molecule binding is not the only way regulator proteins are controlled: OxyR is directly activated by oxidation and phosphorylation of regulatory proteins is a common mechanism
Lactose Operon Review
catabolic, negative regulation
regulatory protein is repressor
effector is the inducer → allolactose (modified lactose)
existence of catabolite activator protein (CAP)
Tryptophan Operon Review
anabolic, negative regulation (making the energy source)
regulatory protein is aporepressor
effector is the co-repressor → tryptophan
autoregulation
L-arabinose Operon Review
catabolic, positive and negative regulation
same regulatory protein acts as both activator and repressor is the regulator with 2 different conformations and corresponding binding sites
effector is the inducer → arabinose
autoregulation of araC and CAP regulation of the operon
Auxiliary Operators
lac-operon has 3 operators:
strongest 01
downstream of CAP binding site 02
upstream of CAP binding site 03
all 3 open means robust transcription of the region
all 3 occupied means transcription is suppressed 1000-fold
if either 02 or 03 are open, it is suppressed 500-fold
both 02 and 03 open suppresses by 20-fold
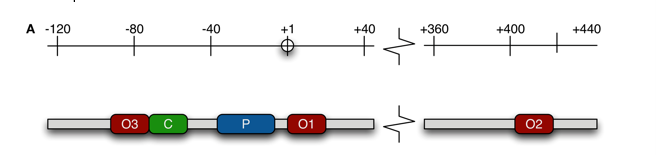
DNA looping prevents _____ from binding to promoter.
RNAP; the repressor binds as a tetramer
presence of auxiliary operators 02 and 03 near the functional operator 01 increases the LOCAL CONCENTRATION of the repressor so it can occupy the functional operator
Other Ways to Regulate Gene Expression
most genes are controlled by protein factors but RNA itself can be regulatory to transcription
differential folding of DNA
riboswitches
alternative sigma factors
Differential Folding of RNA
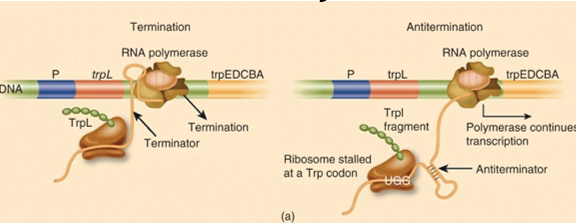
trp operon in bacteria is an example of transcriptional attenuation (putting brake on transcription)
during transcription of the 162-bp leader region of the operon, a domain of the new synthesized RNA transcript can fold to form either an antiterminator (no tryptophan) or terminator (when there is tryptophan) hairpin
antitermination is used as a control mechanism in bacteria to regulate expression of some operons and happens when RNAP reads through
Riboswitches
are structural RNAs that control the gene in an on/off state
typically in 5’ UTR of bacterial mRNAs
most riboswitches can be divided roughly into 2 domains → aptamer and expression platform
can act as metabolite sensors in which the genes controlled by riboswtiches encode proteins involved in the biosynthesis or transport of the metabolite being sensed
with low amounts of lysine, there is read through of termination signals and RNA containing ORF is transcribed
when lysine is abundant there is a structural change when RNA binds to lysine → change in structure that is formed and terminator sequence is now accessible for transcription termination
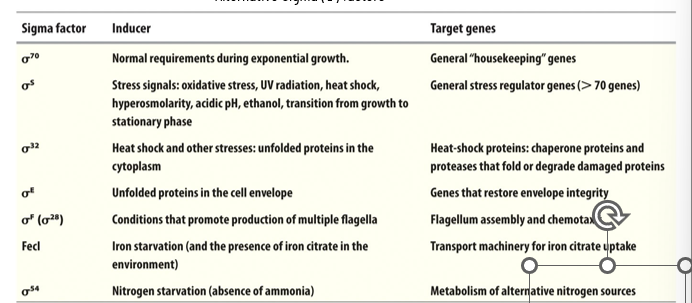
Alternative Sigma Factors
direct holoenzymes to specific promoters - each recognizes a specific promoter sequence
allows specific gene expression for fast initiation under specific conditions
sigma factor 70 is the regular housekeeping but when the organism is under stress we have other sigma factors to help direct RNAP to specific promoters, which are not present under the same conditions every time even if they respond to the same consensus sequence → affects which genes are turned on