Section 3.3: Receptors
1/52
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
53 Terms
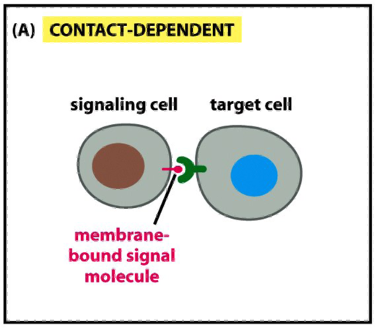
Modes of Communication b/w Cells: Contact Dependent
e.g during development an in immune response
a signaling molecule on the surface of one cell binds to a receptor on a neighboring cell
requires physical contact between cells
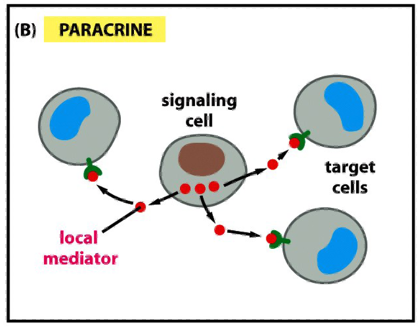
Modes of Communication b/w Cells: Paracrine (autocrine)
signals are released into the extracellular space and act locally on neighboring cells (paracrine)
autocrine: the cell responds to its own secreted signal
eg. cancer cells use this strategy to stimulate survival and proliferation
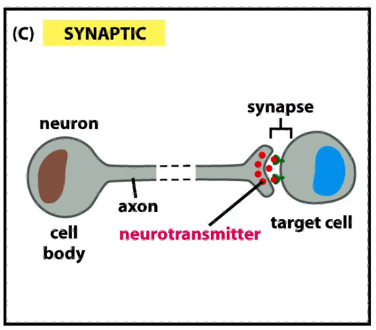
Modes of Communication b/w Cells: Synaptic
neurons transmit signals electrically along their axons
release neurotransmitters at synapses, which are often located far away from the neuronal cell body
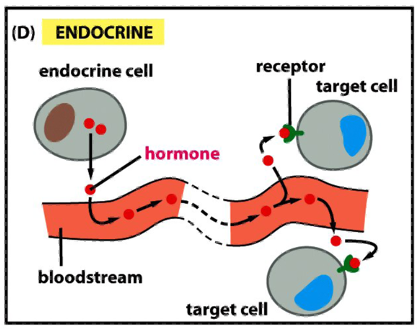
Modes of Communication b/w Cells: Endocrine
endocrine signaling depends on endocrine cells, which secrete hormones into the bloodstream for distribution throughout the body
signaling over long distances makes use of endocrine cells
the same types of signaling molecules are used in paracrine, synaptic and endocrine signaling; the differences lie in the speed and selectivity with whch the signals are delivered to their targets
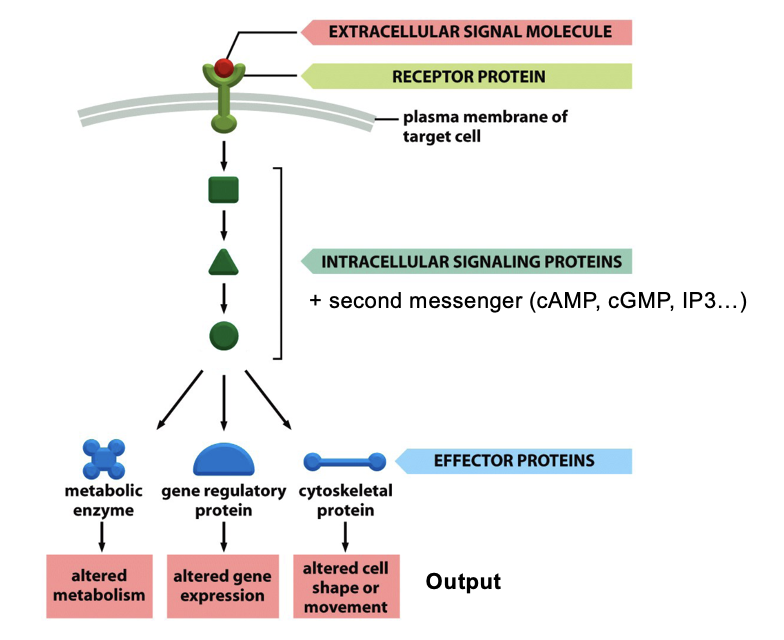
Typical Signaling Cascade
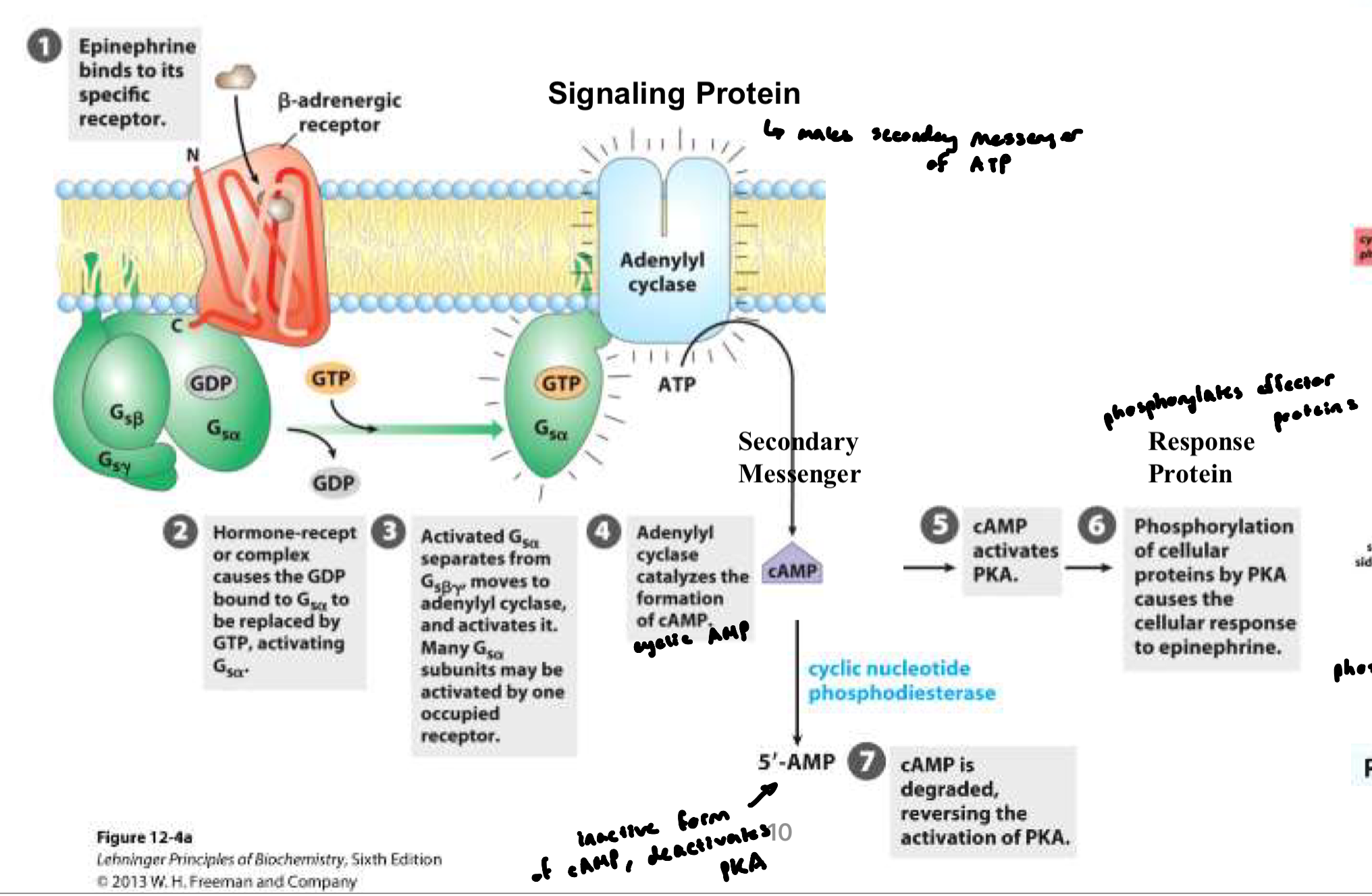
signal molecule binds to a cell-surface receptor
most signal molecules are hydrophilic and cannot cross membrane
activated receptor triggers intracellular signaling pathways (conformational change)
involves a series of signaling proteins and second messengers (cAMP, cGMP, IP3) that relay and amplify the signal
these signaling proteins act on effector proteins, which change cell behavior
GPCR Family
7 TMS
largest family of cell-surface receptor; ~350 GPCRS in humans
bind a variety of ligands: peptides, hormones, growth factors, fatty acids, odorants, light
many GPCRs still have unknown ligands
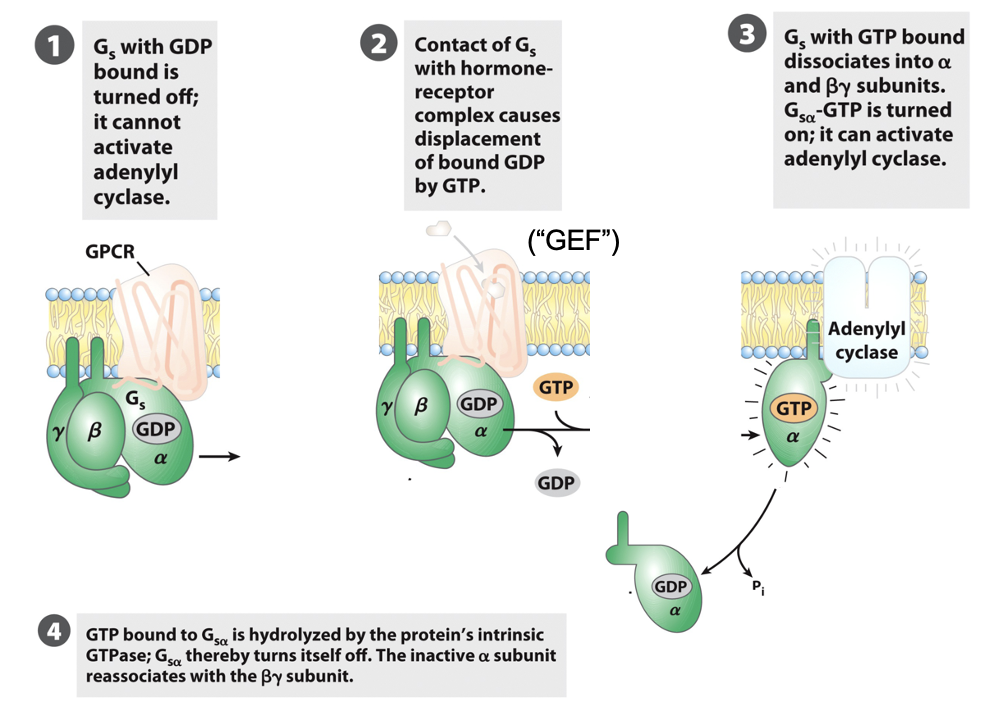
G-Protein Coupled Receptor (GPCR)
membrane receptor bound to a G-protein
GPCRs activate a trimeric G protein on the inner membrane surface
G protein = ⍺, β, γ subunits
Inactive state: G protein has GDP bound to the ⍺ subunit
when ligand binds GPCR (activated), it acts as a GEF
⍺ releases GDP → binds GTP → ⍺ dissociates from βγ → activate target enzymes or ion channels (e.g. in G5, ⍺-GTP activates adenylyl cyclase
G protein remains active until the ⍺ subunit hydrolyzes GTP → reassociates with βγ
G protein has intrinsic GTPase activity stimulated by RGS proteins (regulators of G-protein signaling)
RGS determine how quickly bound GTP is hydrolyzed to GDP and how long G protein remains active
everything stays confined within the bilayer due to the lipid anchor
G-Protein Coupled Receptor (GPCR) FIGURE
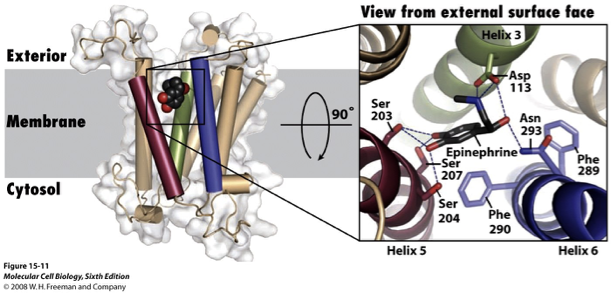
GPCR Example: Adrenaline
when released into blood: increases heart rate, raises blood pressure, opens airways in lungs, boosts blood sugar
mediates mobilization of energy (fight or flight)
β2-adrenergic receptor (β2-AR) is a type of GPCR that responds to adrenaline
epinephrine binds deep within the membrane; the binding site is formed by a.a’s from many TMSs
helices 3,5 and 6 participate in binding
the interaction is stereospecific; 3D orientation of epinephrine is critical for binding (not many things can bind in the pocket and stay there)
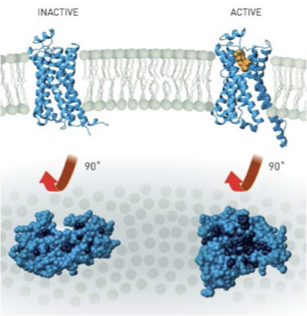
β2-AR: Active vs Inactive
inactive state: bound to carazolol (inverse agonist or antagonist)
active state: part of the β2AR-G’s complex
TM6 moves outward to allow G-protein binding
TM5 and TM3 also shift subtly to transmit the signal
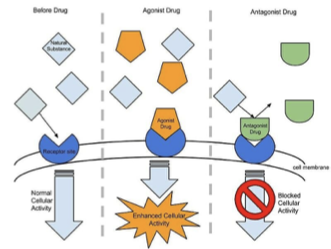
Agonist
binds GPCR and stabilizes active form → activates G-protein
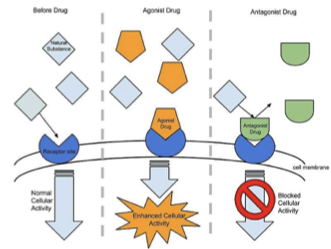
Inverse Agonist
stabilizes the inactive form of the receptor
Antagonist
blocks receptor activation by preventing the conformational change that would activate the G protein
Adenylate Cyclase Pathway
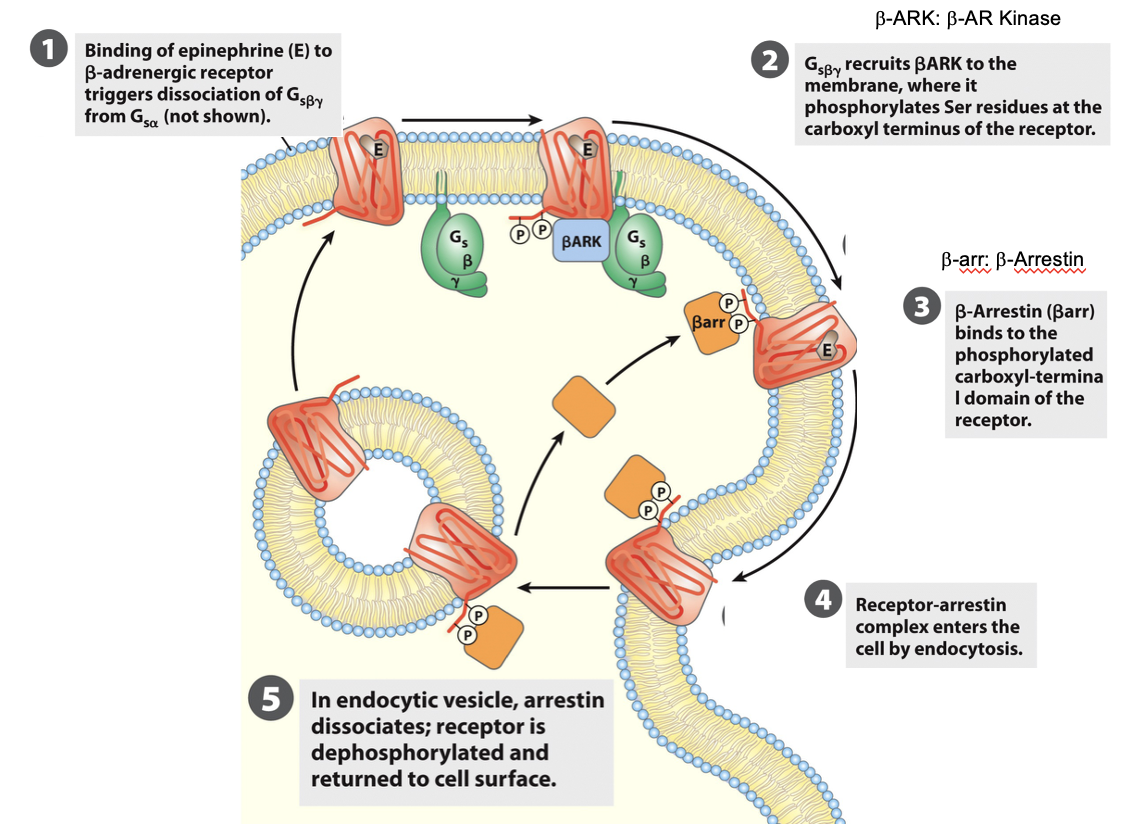
Desensitization: β2 Arrestin
after prolonged stimulation, β-arrestin binds to the receptor → prevents further G protein activation → receptor desensesitization
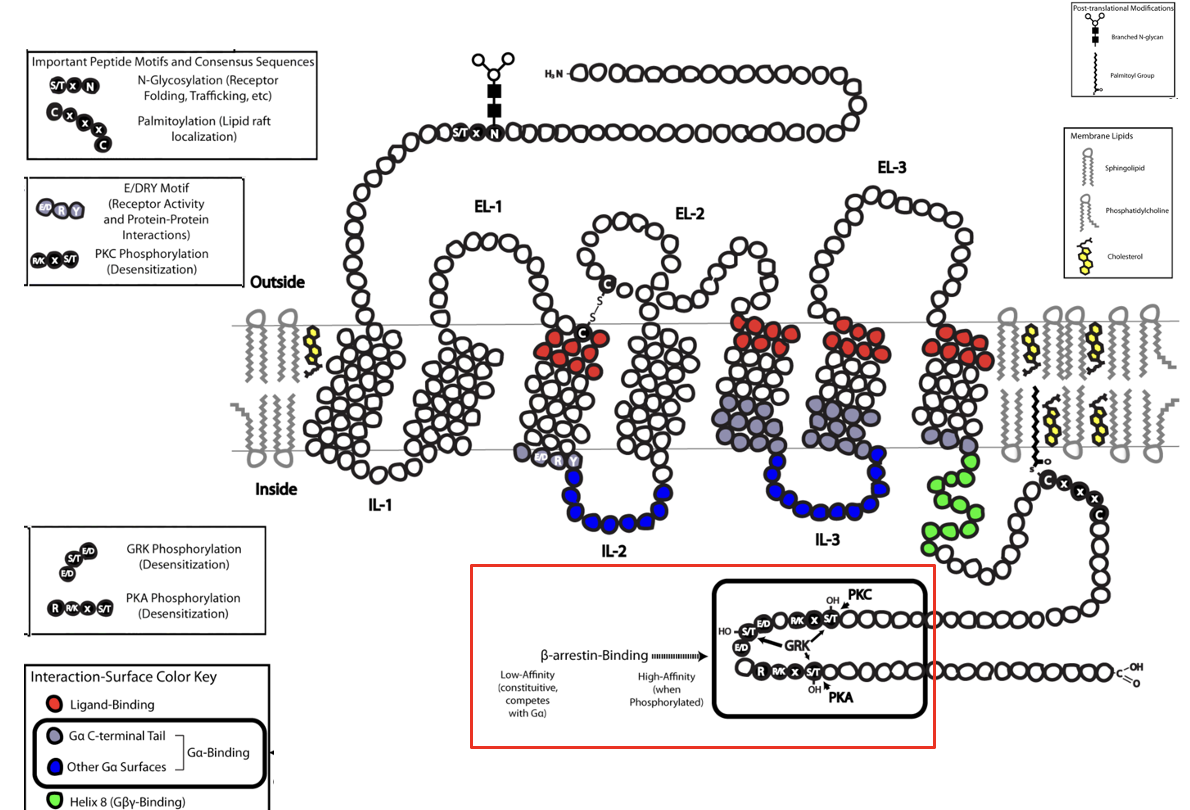
Desensitization to Adrenaline
when epineprine is present continuously, β-adrenergic receptors respond less over time (desensitization, leading to a reduced cellular response)
eg. chronic stress
Key proteins:
β-adrenergic receptor kinase (βARK): phosphorylates receptor on C-terminal
β-arrestin: binds phosphorylated receptor → prevents further G-protein activation
Epinephrine and Synthetic Analogs
epinephrine binds β-adrenergic receptors; affinity is measured as dissociation constant (Kd) of receptor-ligand complex
synthetic analogs: chemically modified versions of epinephrine that can either mimic or block its action
isoproterenol: synthetic agonist with higher affinity than epinephrine (strongly activates β-receptors)
propranolol: synthetic antagonist (beta blocker), extremely high affinity → blocks receptor activation
Receptor Ligand Binding Interaction
rate of formation of RL complex: kon [R] [L]
rate of dissociation of RL complex: koff [RL]
at equilibrium: rate of formation = rate of dissociation
Kd = koff / kon = ([R] [L])/[RL]
Kd: when 50% of receptor is bound to ligand
low Kd → high affinity (less ligand needed to occupy 50% of receptors)
![<ul><li><p>rate of formation of RL complex: k<sub>on</sub> [R] [L]</p></li><li><p>rate of dissociation of RL complex: k<sub>off</sub> [RL]</p></li><li><p>at equilibrium: rate of formation = rate of dissociation</p></li><li><p>Kd = k<sub>off </sub>/ k<sub>on</sub> = ([R] [L])/[RL]</p><ul><li><p>Kd: when 50% of receptor is bound to ligand</p></li><li><p>low K<sub>d</sub> → high affinity (less ligand needed to occupy 50% of receptors)</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/d998120b-55ae-437c-a49f-de8887192563.png)
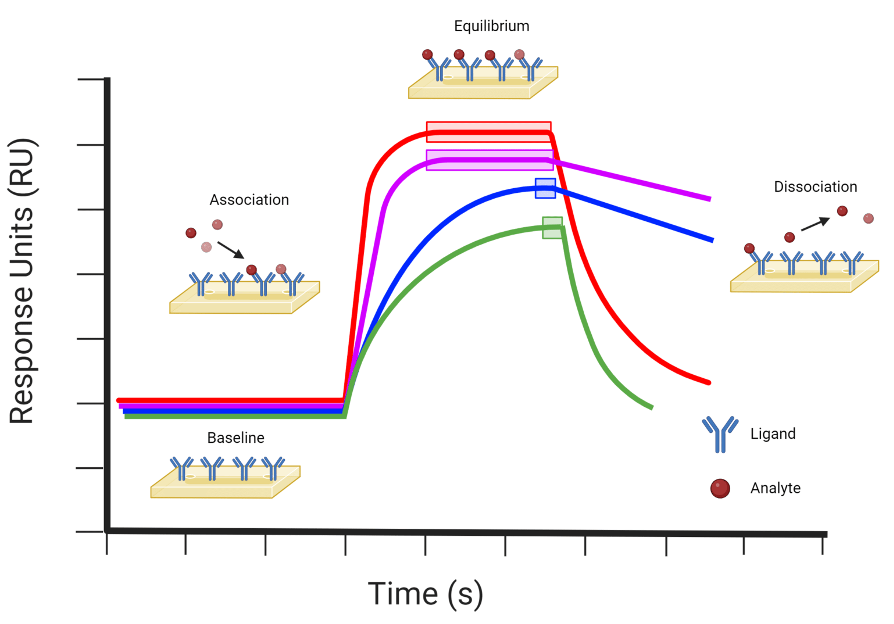
Receptor Ligand Interaction Experiment: Surface Plasmon Resonance (SPR)
technique used to measure binding interactions in real time w/o labeling the ligand or receptor
produces a sensorgram, showing response (binding) vs time
baseline: before ligand is introduced → no binding
association phase: ligand binds receptor → signal increases
equilibrium phase: rate of binding = rate of dissociation → plateau in signal
dissociation phase: ligand removed → signal decreases
Receptor Ligand Interaction Experiment: Surface Plasmon Resonance (SPR) FIGURE
Red: fast association, fast dissociation → transient binding.
Purple: fast association, slow dissociation → strong/stable binding.
Blue: slow association, slow dissociation → gradual, stable binding.
Green: slow association, fast dissociation → weak, transient binding.
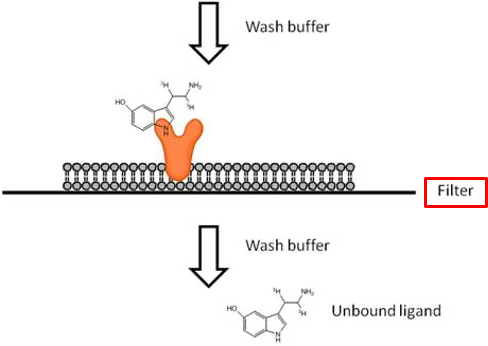
Receptor Ligand Interaction - Experimentation
isolate cells (or membranes) containing the receptors. Place onto the filter
prepare saturating amounts of ligand molecules (eg. radioactive or fluorescent)
pass the mixture through the filter (pore small enough to retain cells or membranes)
wash away unbound ligand molecules
measure bound radioactivity (the sum of specific + non specific binding)
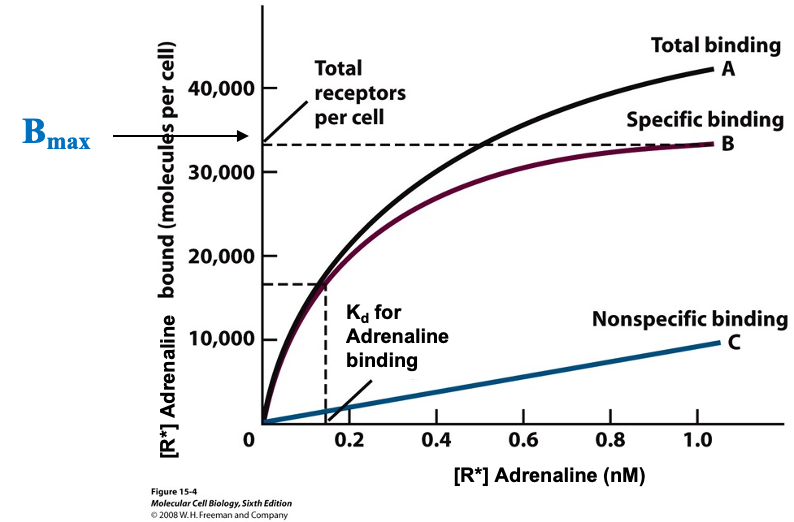
Binding Assay- Typical Curve
cells w/ receptors: 1000-50000 copies per cell
cells were incubated for 1 hour at 4ºC with radioactively labeled adrenaline
assume no endocytosis of the cell is taking place
curve A: adrenaline bound to receptors and non specifically bound (never reach a plateau)
curve B: difference between A and C (ideal)
this type of curve allows determination of receptor number (Bmax) and Kd
CFTR (-/-) Mice are Resistant to Cholera Toxin
people who are carriers of cystic mutation (CFTR +/-) may receive a survival advantage in diseases that cause massive salt and water loss (eg. cholera)
CFTR is the Cl- channel that cholera toxin hijacks to cause secretory diarrhea
cholera forces CFTR to stay permanently open
Mouse Experiment:
CFTR (-/-): cannot secrete chloride → cannot develop cholera diarrhea
CF (+/-): reduced CFTR activity → less activity than normal mice
WT (+/+): full CFTR function → strong diarrhea response to cholera
Regulation of CFTR (ABCC7) by PKA
CFTR carries a regulatory domain (R-domain) that is phosphorylated and regulates transporter activity
phosphorylated = open; dephosphorylated = blocks channel gate (no Cl- flow)
β-adrenergic signaling increases cAMP, PKA is activated and phosphorylates the R domain
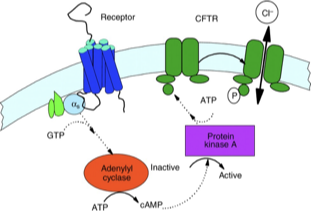
cAMP
Cyclic Adenosine Monophosphate
intracellular second messenger molecule involved in many cell signaling pathways
relaying signals from hormones like adrenaline to activate enzymes, open channels, and regulate genes
Vibrio Cholerae
Vibrio cholerae is the bacterium that causes cholera; to cause disease i must deliver cholera toxin into intestinal epithelial cells
the cholera bacterium has a large secretion system that spans the inner membrane, periplasm and outer membrane
this apparatus is ATP powered
function is to export cholera toxin out of the bacteria and into the environment/host
Pre-Ctx A/B
precursor forms of cholera toxin, subunits A and B
include a signal peptide that directs them thru the Sec secretion system
cannot fold in cytosol (becomes stuck thru Sec pore → folding occurs after protein is in periplasm → delivered to secretion apparatus (the one than spans the multiple membranes and ATP powered)
Cell Penetration and Action of Cholera Toxin Part 1
cholera toxin (CT) = AB5 toxin (6 subunits)
CT binds to GM1 glycosphingolipid on intestinal epithelial cell surface → toxin is endocytosed in retrograde direction (endosome → Golgi → ER)
CtxA contains a KDEL sequence (guides direction to ER instead of lysosome)
in the ER, cholera toxin mimics a misfolded protein
protein disulphide isomerase (PDI) breaks the disulfide bond that links CtxA and B
Cell Penetration and Action of Cholera Toxin Part 2
once freed, CtxA1 is recognized as misfolded and transported to cytosol via Sec61 complex → most of CtxA1 is degraded by proteasome
remaining fragment is enzymatically active → transfers ADP-ribose moiety of NAD+ to G-⍺ subunit, inactivates GTPase activity→ Gs⍺ is always active
always active Gs⍺ → increased production of cAMP (activated adenyl cyclase) → activates protein kinase A (PKA) → CFTR phosphorylated and permanently open → massive Cl- efflux → Na+ and water follow → diarrhea
Cell Penetration and Action of Cholera Toxin FIGURE
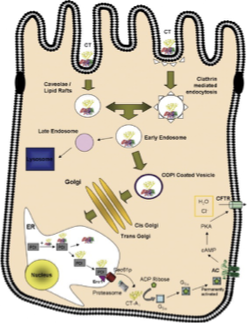
Another Cholera Toxin Figure

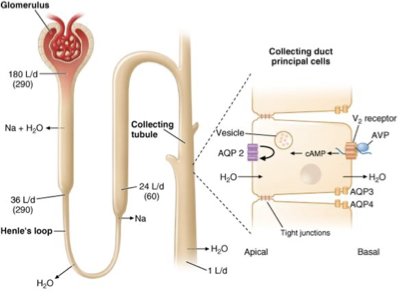
Anti-Diuretic Hormone (ADH): Background
9 a.a peptide
in 24H, kidneys produce ~170L of primary urine, but extensive water reabsorption controls it to 1L being excreted
the recycling machinery is possible b/c of aquaporins (AQPs) (millions in a single kidney)
ADH (aka vasopressin, AVP) promotes the insertion of AQP2 channels to CM of renal tubular cells → increasing water reabsorption from urine
ADH deficiency leads to diabetes insipidus (excessive urine production)
ADH: Vasopressin Receptor Signaling (V2R)
the binding of ADH to its receptor V2R activates a G-protein coupled signaling cascade
AVP binding → activation of V2R → activates adenylate cyclase → increased cAMP levels→ activates PKA → triggers exocytosis of vesicles containing AQP2
increased AQP2 at CM = enhanced water reabsorbion
ADH: Vasopressin Receptor Signaling (V2R)
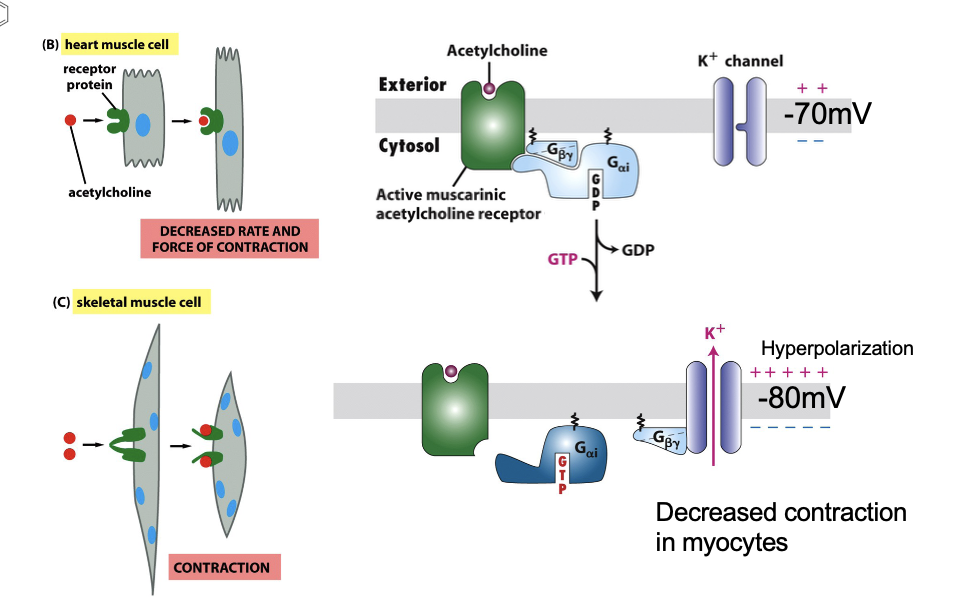
Muscarinic Receptor (GPCR) Background
muscarine: acetylcholine analog
binds more strongly to muscarinic acetylcholine (mAChR) than acetylcholine
mAChR is a GPCR, coupled to G⍺i protein
atropine antidote antagonist
Muscarinic Receptor (GPCR): Mechanism of Action in Heart Muscle
muscarine binds mAChR
G⍺i dissociates from Gβγ upon GTP binding → K+ channels open → K+ efflux → hyperpolarization (more negative membrane potential) → keeps voltage-gated Ca2+ channels closed → reduces frequency of heart muscle contraction
Muscarinic Receptor (GPCR): Termination of Signaling
G⍺i hydrolyzes GTP→ GDP
G⍺i-GDP recombines with Gβγ → channel closes → normal Vm restored
Muscarinic vs Nicotinic Receptors
Nicotinic ACh receptor: ligand-gated ion channel → fast depolarization → muscular contraction
Muscarinic ACh receptor: GPCR → slower, indirect effect thru G protein → muscular relaxation
Muscarinic vs Nicotinic Receptors FIGURE
Light Receptor - Rhodopsin: Anatomy of Retina
Rods: responsible for high resolution and night vision
Cones: color vision, 3 subtypes
rods and cones form synpases with interconnecting neurons, which relay signals to ganglion cells → optic nerve → visual cortex
Light Receptor - Rhodopsin: Rod Cell Structure
outer segment: contains ~1000 stacked discs with rhodopsin
discs are not connected to PM
inner segment: cell body with nucleus and organelles
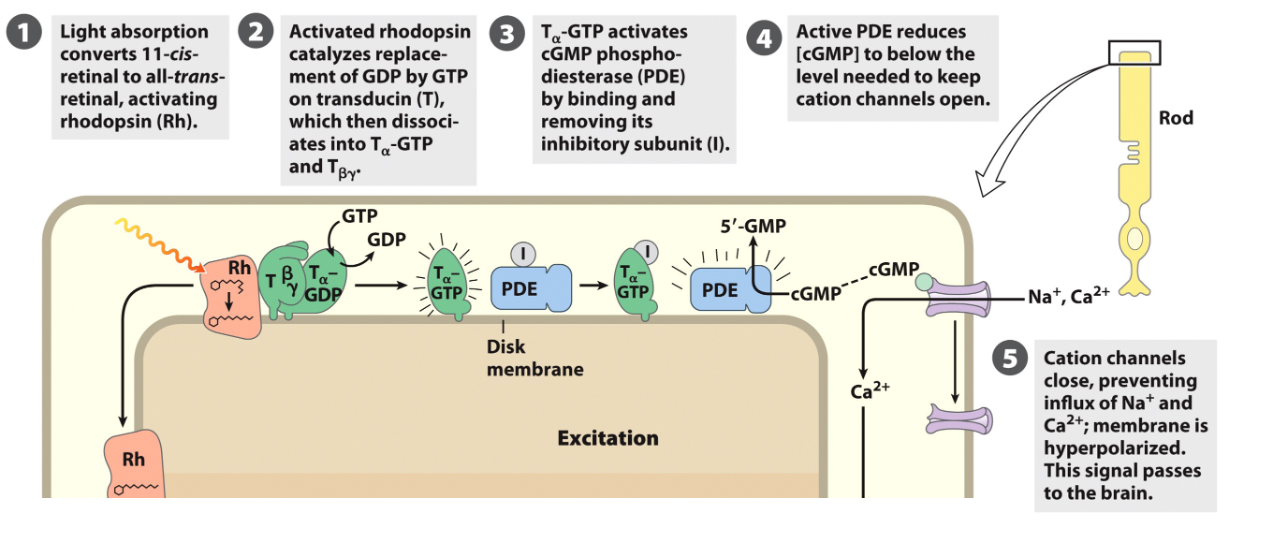
Rdodopsin: GPCR Light Receptor/Phototransduction Cycle
rhodopsin in the disc membrane contains a chromophore (11- cis retinal)
GPCR activated by a photon
photon → 11 cis isomerizes to all-trans retinal
rhodopsin undergoes conformational change → meta rhodhopsin II (active opsin)
meta-rhodopsin II activates transducin (Gt) by promoting GTP binding to G⍺t
G⍺t-GTP then interacts with phosphodiersterase (PDE γ subunits)
Phototransduction Cycle FIGURE
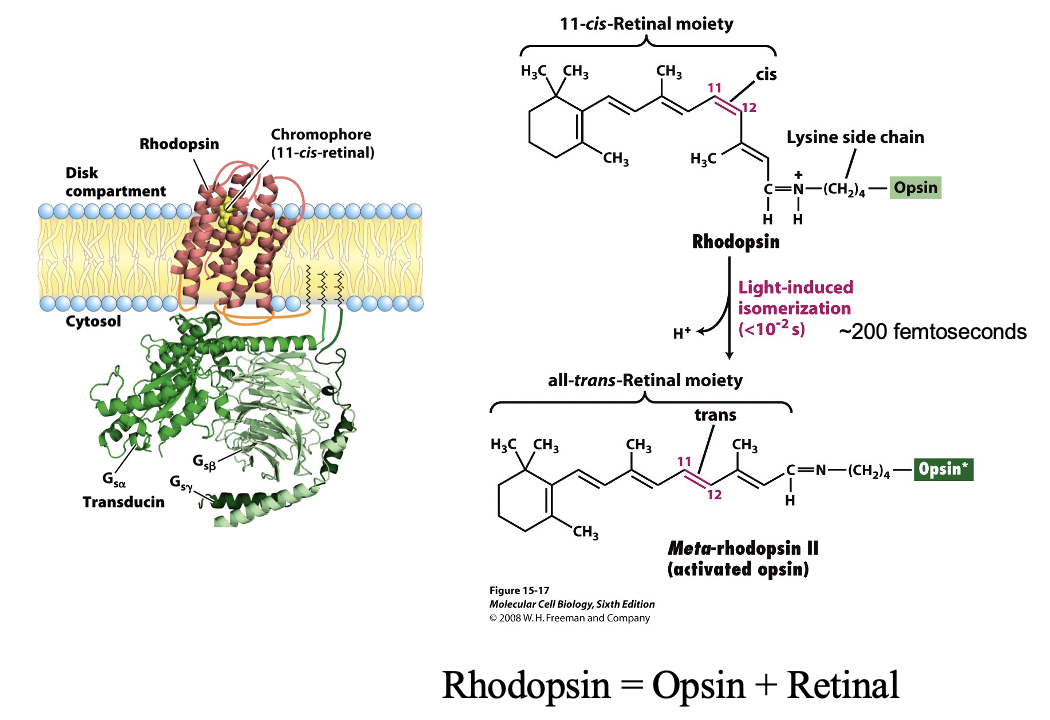
Rhodopsin: GPCR Light Receptor: Chromophore Recycling
all trans retinal dissociates from opsin
enzymes convert it back to 11-cis retinal
rebinds opsin → ready for next photon
Rhodopsin Figure
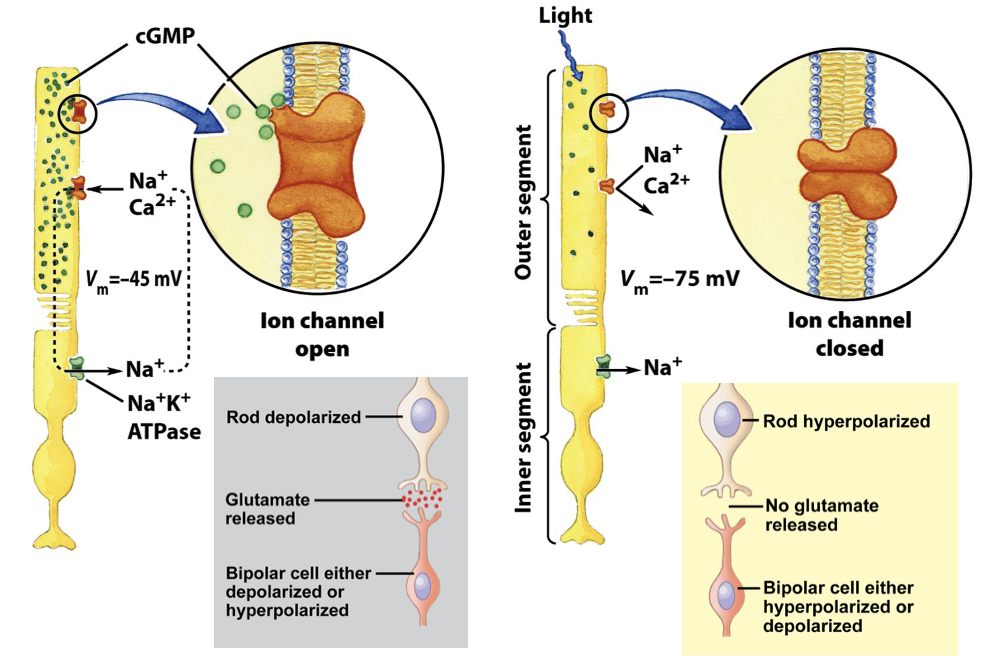
Rhodopsin: GPCR Light Receptor: cGMP gated Ion Channel in Rod Cells
activation of PDE → PDE hydrolyzes cGMP → GMP → [cGMP] decreases
Na+/Ca2+ channels in the rod outer segment require cGMP to stay open
low [cGMP] → channels close
rod cell hyperpolarizes → membrane potential becomes more negative
hyperpolarization reduces neurotransmitter release
light essentially inhibits the electrical signal
ATP in the inner segment of the rod powers the Na+/K+ ATPase, creates a transmembrane electrical potential
cGMP gated Ion Channel in Rod Cells FIGURE
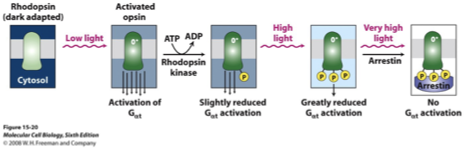
Adaptation/Desensitization of Phototransduction Pathway
Opsin phosphorylation: light-activated opsin can be phosphorylated by a rhodopsin kinase
more light → more opsin in active state → more phosphorylation
Effect on G protein activation: phosphorylated opsin is less able to activate G⍺t (transducin)
in a bright light, a larger amount of light is needed to generate the same signal (light adaptation)
Arrestin binding: at very high light levels, arrestin binds fully phosphorylated opsin
opsin-arrestin complex cannot activate G⍺t at all → phototransduction temporarily halted
protects cell from overstimulation and saturation
Adaptation/Desensitization of Phototransduction Pathway
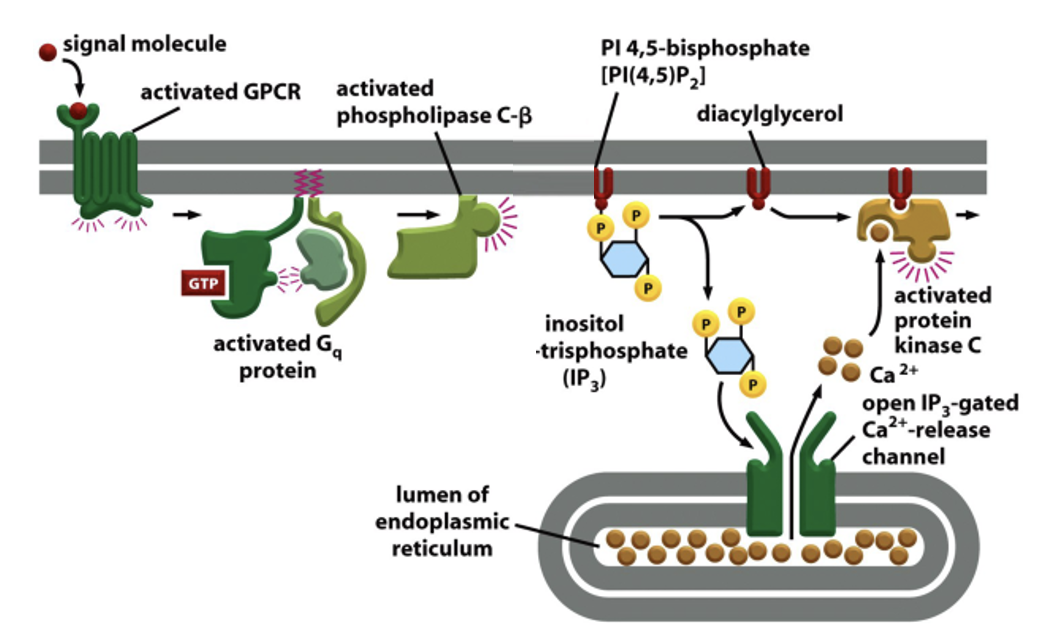
GPCR linked to the IP3 Pathway
certain GPCRs activate phospholipase C (PLC)
PLC cleaves PIP2 in the PM to generate inositol 1,4,5 trisphosphate (IP3) cytosolic messenger, and DAG membrane bound messenger
at ER membrane, Ca2+ opens IP3 gated Ca2+ release channels (IP3 receptor)
Ca2+ stored in the ER quickly rises in cytosol
DAG stays in PM; tgt w/ phosphatidylserine and Ca2+, helps activate protein kinase C (PKC)
PKC phosphorylaes target proteins
GPCR linked to the IP3 Pathway: Termination of the Signal
IP3 dephosphorylated → inactivated (by specific lipid phosphatases)
IP3 phosphorylated → form IP4 (by specific lipid kinases)
Ca2+ that enters the cytosol is rapidly pumped out, mainly to exterior of the cell
GPCR linked to the IP3 Pathway: Figure
Sweet Receptor and IP3 Pathway
sweet receptor is a GPCR on taste receptor cells
activation occurs when a sweet molecule binds
inositol 1,4,5 trisphophate diffuses thru the cytosol and releases Ca2+ from the ER by binding to an opening IP3-gated Ca2+ release channels