Quantitative real time PCR and detection
1/12
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
13 Terms
PCR
Polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it to a large enough amount to study in detail. it is used to identify (qualification) and amplify a sequence of DNA.
Real time PCR is done also for quantification use and for other aims:
Quantification of gene expression levels
Quantification of DNA (Telomere assays, mtDNA content, copy number)
Virus titre (quantification of very low numbers)
Bacteria/protozoa identification and antibiotic resistance
Can be used as a quantitative step as part of another technique e.g. ChIP and DNA Methylation
PCR workflow
Preparation of the sample with homogenization, weighting, or enrichment (putting the sample in an enrichment culture and allowing it to grow). The sample needs to be representative of the medium it comes from.
Reaction mix.
PCR setup. Reaction mix is added with the Taq polymerase equals to master mix. Depending on the application, a proofreading Taq polymerase is used (ex. if you need the exact copy of the gene). Different polymerases amplify at around 78°C but some work at 65°C, some are most indicated to amplify longer or shorter fragments, work best with different GC% (most work best with 50/50 GC, but for example, plasmodium has a 85% GC, so we need specific polymerases).
denaturation at 95°C, permitting the opening of the elix. It lasts a few seconds.
annealing at 65°C of the primers forward and reverse to the region that is going to be amplified. Primers should be at least 18 nucleotides long (the lenght makes it more specific). It lasts 30 seconds at most.
extension at 72°C, the polymerase builds the remaining of the sequence next to the primers. It lasts 1 minute for 1kb.
Reaction mix.
Primers specific to target: 18–24bp, produce product 50 – 150 bp in length (200bp does work), product ~50% GC and avoid GC stretches and repetitive sequences, check for primer dimers especially for SYBR green assays, F and R primers annealing temp within 5˚C, in separate exons or spanning exon boundaries
Probes: Pre-designed assay available (TaqMan probes or sybr based assays)
Template DNA: 50-100ng/uL
Reaction buffer (Tris HCl, ammonium ions, KCl, Mg ions): provides the ionic strength and buffering capacity for the reaction
MgCl2
dNTPs in equimolar ratios
DNA polymerase: 1/2 unit/25 uL reaction
What’s Wrong With Agarose Gels?
Low sensitivity
Low resolution
Non-automated
Size-based discrimination only
Results are not expressed as numbers, so it is based on personal evaluation
Ethidium bromide staining is not very quantitative and toxic
sybr green
Dye binds minor groove of all DNA
Fluorescence much stronger when bound
More product more dye bound
Cheap, not specific
At early stage check product on gel and perform dissociation analysis (melt curve)
probe
Dye and quencher either end of probe
Nuclease activity of polymerase removes dye and quencher during PCR
More product produced, more fluorescence
No melt curve
Allows multiplexing
Absolute quantification.
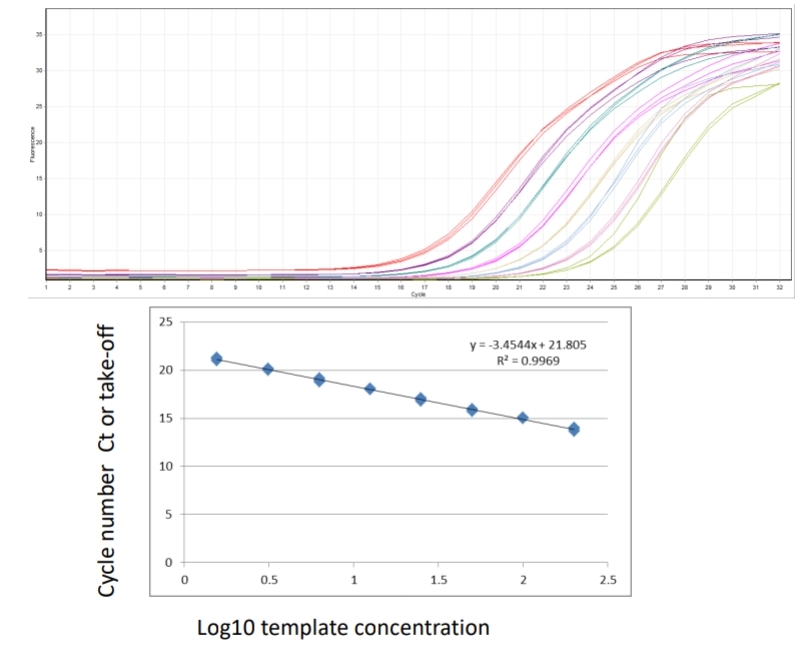
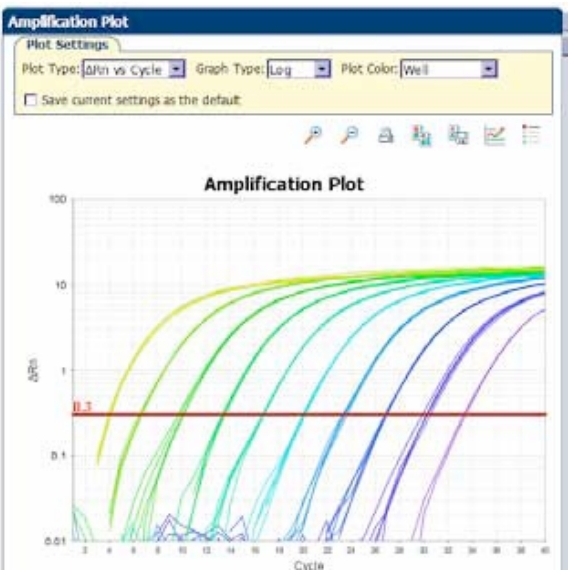
Samples calculated against a standard curve of known concentration. Allows quantification as copy number, takes some variability into account, requires more reagents and a known standard. Plasmid DNA containing product, no RT step so efficiencies may be different, in-vitro transcription to generate specific mRNA – RT, costly. What is a standard curve and why is it necessary? Real time PCR conducted across a series of serially diluted template/samples tells you what the dynamic (linear) range of the assay is and allows calculation of efficiency.
Relative quantification.
Comparative quantification between the gene of interest and an housekeeping gene. Housekeeping gene(s). Accounts for pipetting error and differences in RT efficiencies between samples, constantly expressed at a stable level. Put the same starting RNA in all RT-reactions when samples are to be compared, but doesn’t take account for massive differences in starting material.
TaqMan probes
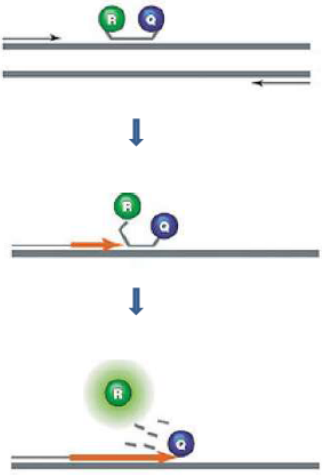
In TaqMan Probes (hydrolysis probes) when intact, the fluorescence of the reporter is quenched due to its proximity to the quencher. Probe hybridizes to the target, dsDNA-specific 5'->3' exonuclease activity of Taq or Tth cleaves off the reporter. Reporter is separated from the quencher → Fluorescent signal proportional to the amount of amplified product in the sample.
Molecular Beacons
Molecular Beacons are hairpin probes composed of a (25-40 nt) nucleotide base paired stem and a target specific nucleotide loop. The loop consists of target specific nucleotide (probe) sequences (15-30 nt). A fluorescent moiety (reporter) is attached to 5' end and a quencher moiety is attached to 3'end. The stem keeps both the moieties in close proximity so that fluorescence is quenched.
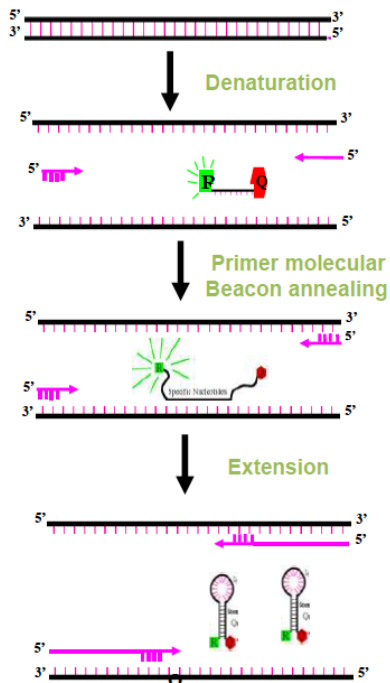
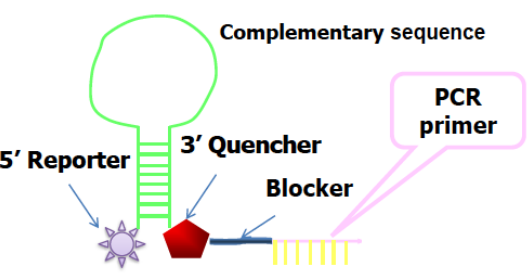
Scorpions probe
The loop of the Scorpions probe includes a sequence that is complementary to an internal portion of the sequence it primes. During the first amplification cycle, the Scorpions primer is extended, and the sequence complementary to the loop sequence is generated. After subsequent denaturation and annealing, the loop of the Scorpions probe hybridizes to the internal target sequence, and the reporter is separated from the quencher. The resulting fluorescent signal is proportional to the amount of amplified product in the sample. The Scorpions probe contains a PCR blocker just 3' of the quencher to prevent read-through during the extension of the opposite strand.
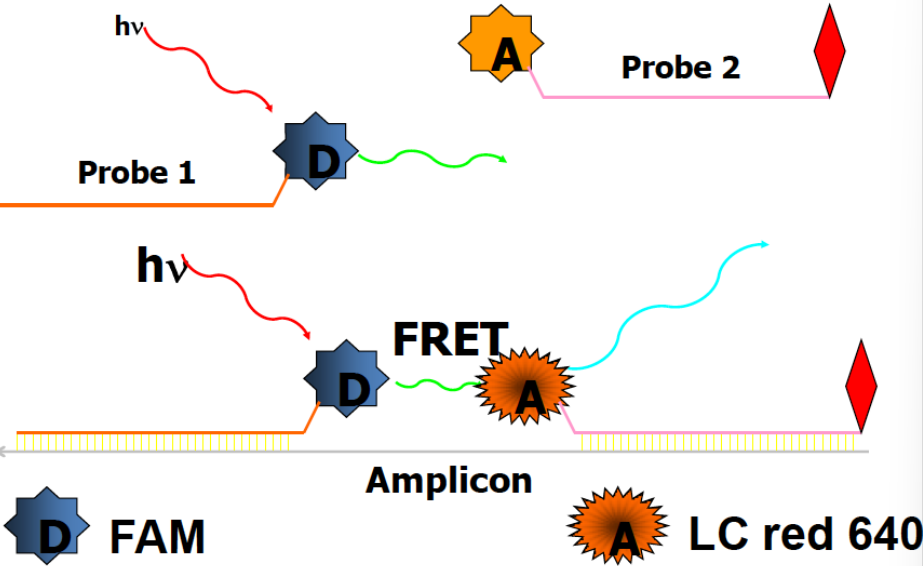
Hybridization Probes.
These assays use two sequence-specific oligonucleotide probes in addition to two sequence specific primers. The two probes are designed to bind to adjacent sequences in the target. The probes are labeled with a pair of dyes that can engage in FRET. The donor dye is attached to the 3′ end of the first probe, while the acceptor dye is attached to the 5' end of the second probe. During real-time PCR, excitation is performed at a wavelength specific to the donor dye, and the reaction is monitored at the emission wavelength of the acceptor dye. At the annealing step, the probes hybridize to their target sequences in a head-to-tail arrangement. This brings the donor and acceptor dyes into proximity, allowing FRET to occur. The increase in PCR product is proportional to amount of fluorescence