Chapter 7: Enzymes
1/106
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
107 Terms
catalyst
change the rate of a rxn by lowering Ea
doesn’t change during a rxn
incr rxn rates without being used up, so does not alter equilibrium
enzyme
a substance produced by a living organism that is a catalyst for a biochemical rxn
a vast majority of biological catalysts are globular proteins (but a few are RNAs)
does induced fit during the rxn with the substrate
substrate
the ligand for an enzymatic rxn that gets changed by the rxn
product
the result of an enzymatic rxn
active site
the location on an enzyme where a rxn occurs
very specific for substrates
3 main ways enzymes are used
diagnosis and prognosis of diseases (by measuring the amount of enzyme in the body)
as analytical reagents in the measurement of nonenzyme substances (can convert non-detectable molecules to detectable ones like drugs, hormones, etc)
as therapeutic agents (ex to reopen blood vessels)
mild inflammatory conditions release ____ enzymes
cytoplasmic enzymes
necrotic conditions release ____ enzymes
mitochondrial enzymes
enzymes can incr rxn rates by up to _____ times as fast
x10^19
than the uncatalyzed rxn
rate enhancement
shows how much an enzyme increases the rxn rate
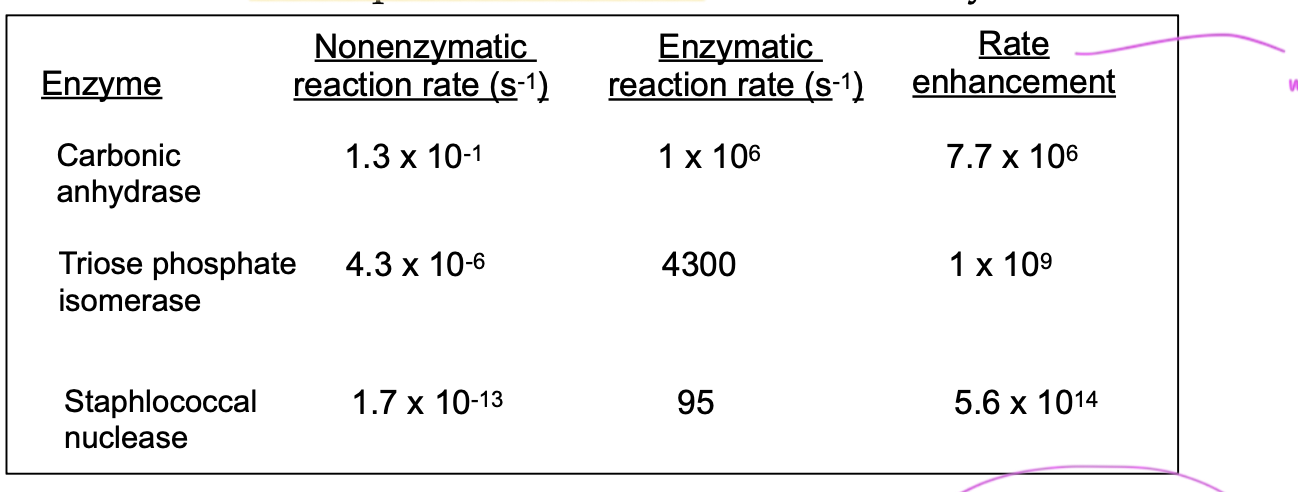
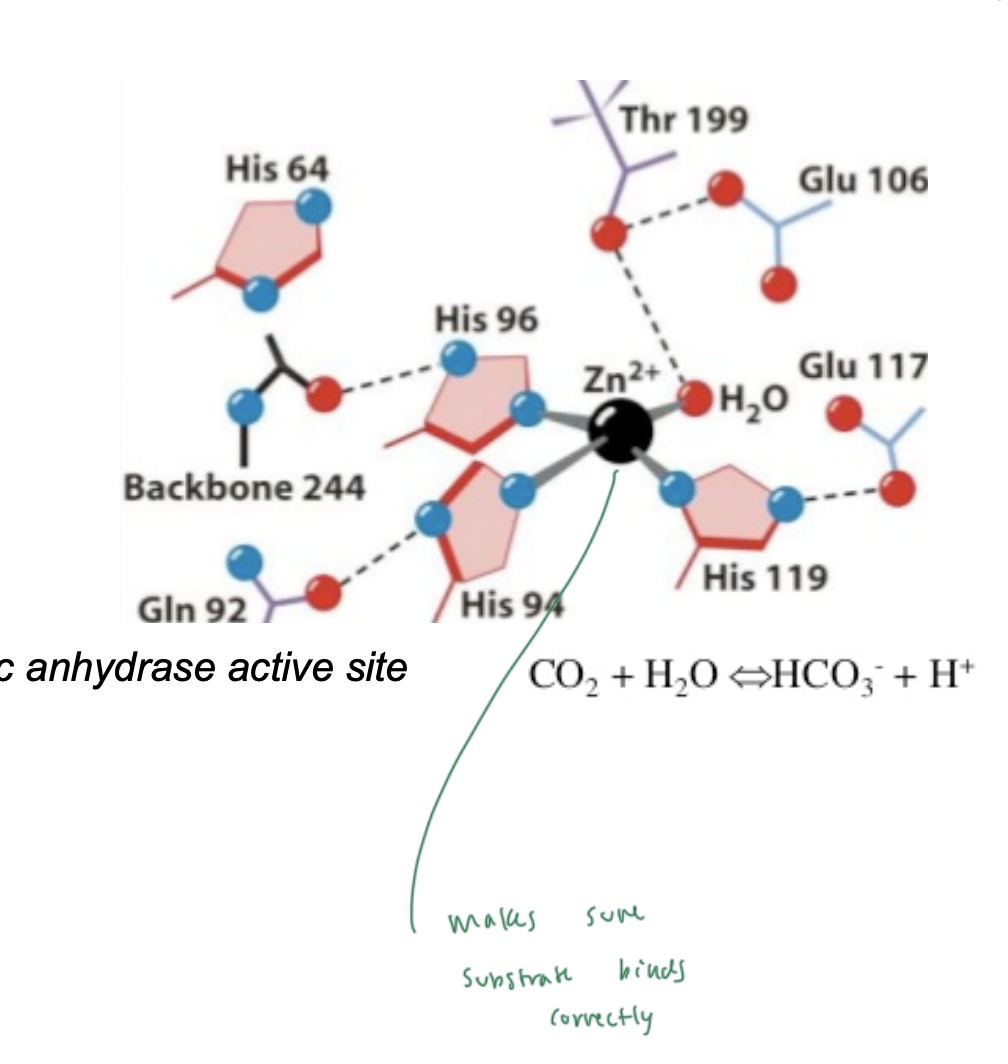
carbonic anhydrase
enzyme that does the carbon dioxide rxn in the lungs
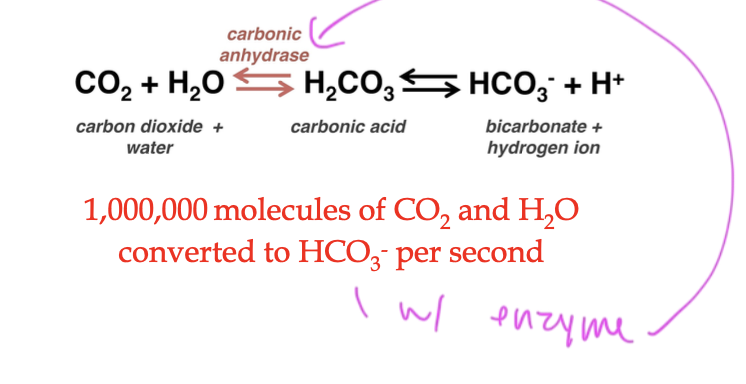
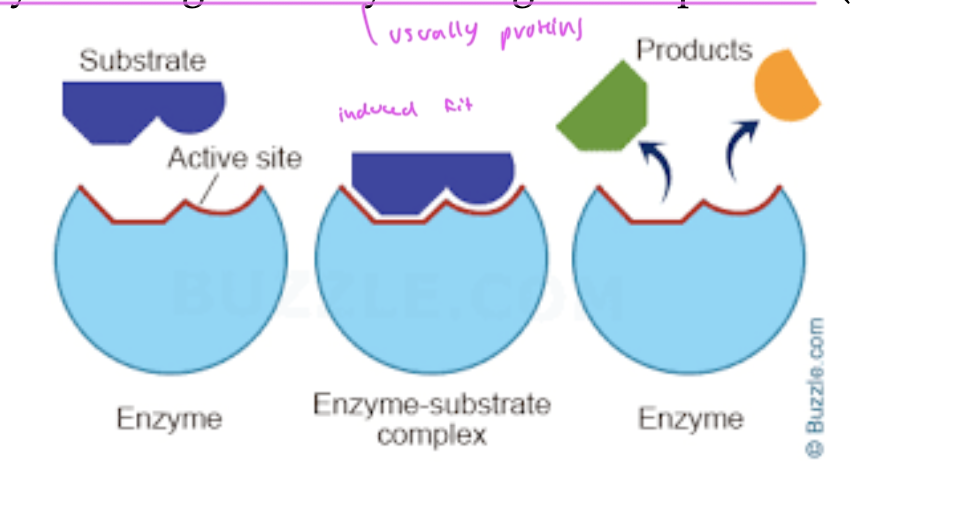
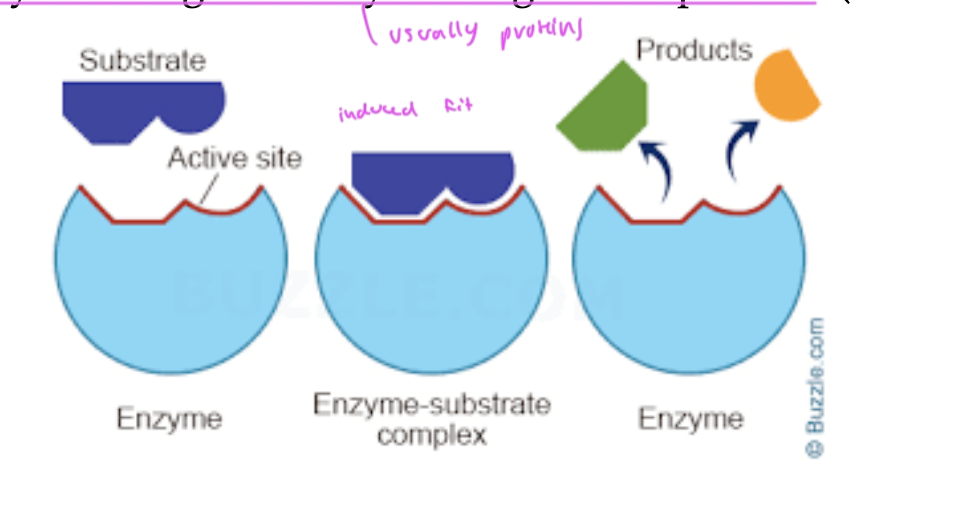
steps of an ezymatic rxn
enzyme interacts with the substrate, does induced fit
now you have an enzyme-substrate complex
a product starts being formed
the product (changed substrate) gets released from the enzyme

enzymes act in the ____ and ____ rxns
forward and backward
they speed up both directions of the rxn, helping the rxn reach equilibrium
what types of interactions drive active site binding affinity?
shape
charge
hydrophobics
any molecular complementarity that is NOT permanent (like covalent bonding)
factors regulating enzyme activity
pH
temp
enzyme conc
substrate conc
cofactor and coenzyme conc
1/3 of enzymes are
metalloenzymes
require metal ions as cofactors/coenzymes
holoenzyme
a apoenzyme (just an enzyme) with its cofactor/coenzyme/metal ion
a functional complex
the enzyme is not functional without its cofactor
coenzymes are often
vitamins that are essential for our diet
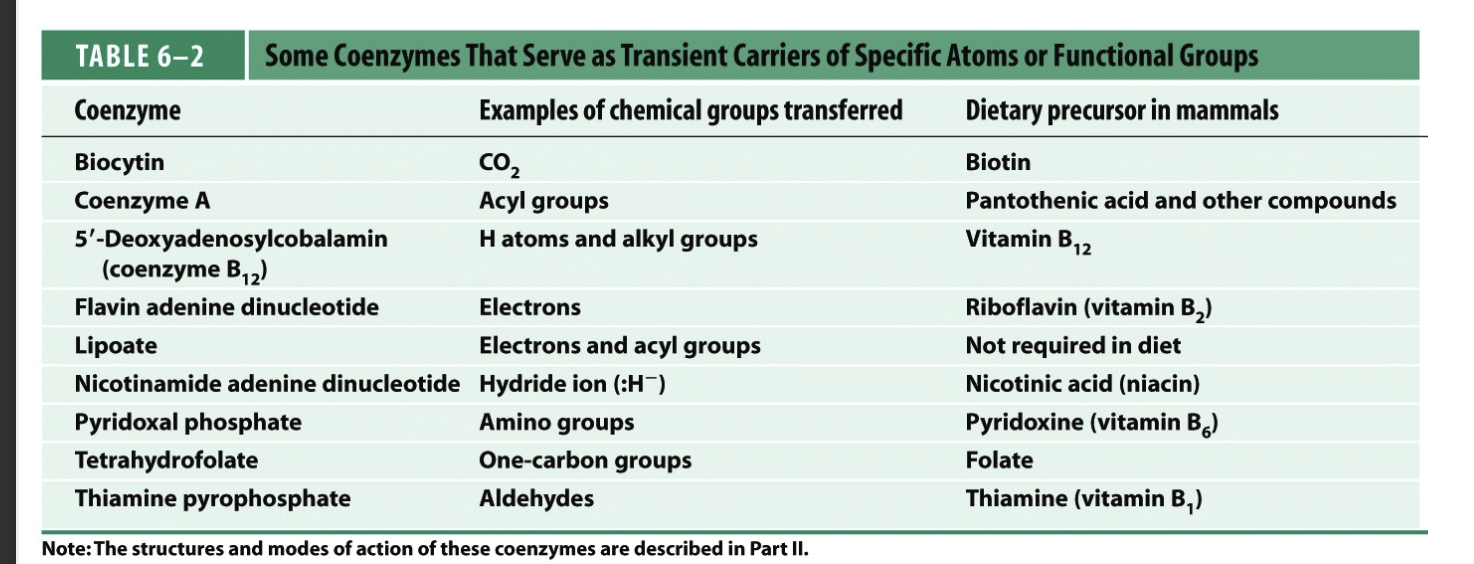
some common cofactors
often metal cations

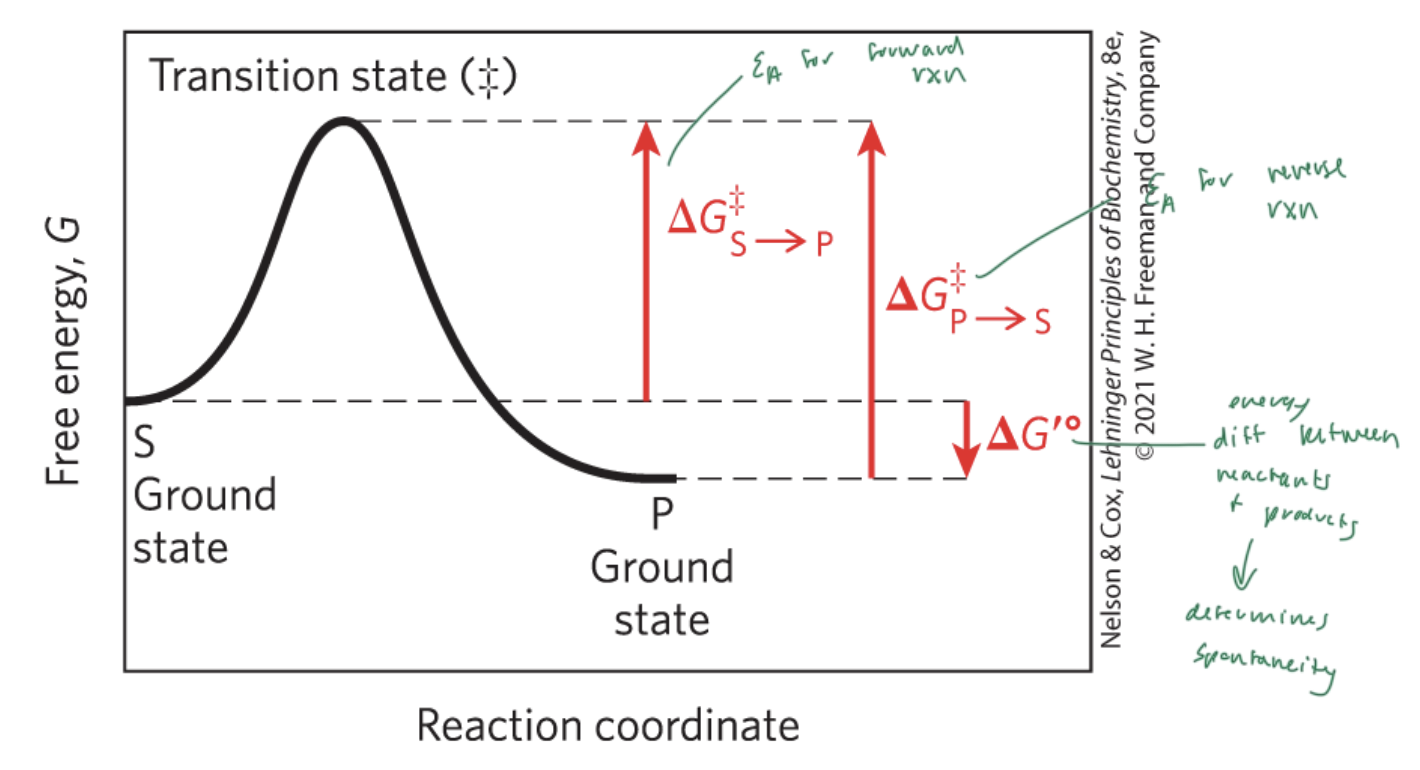
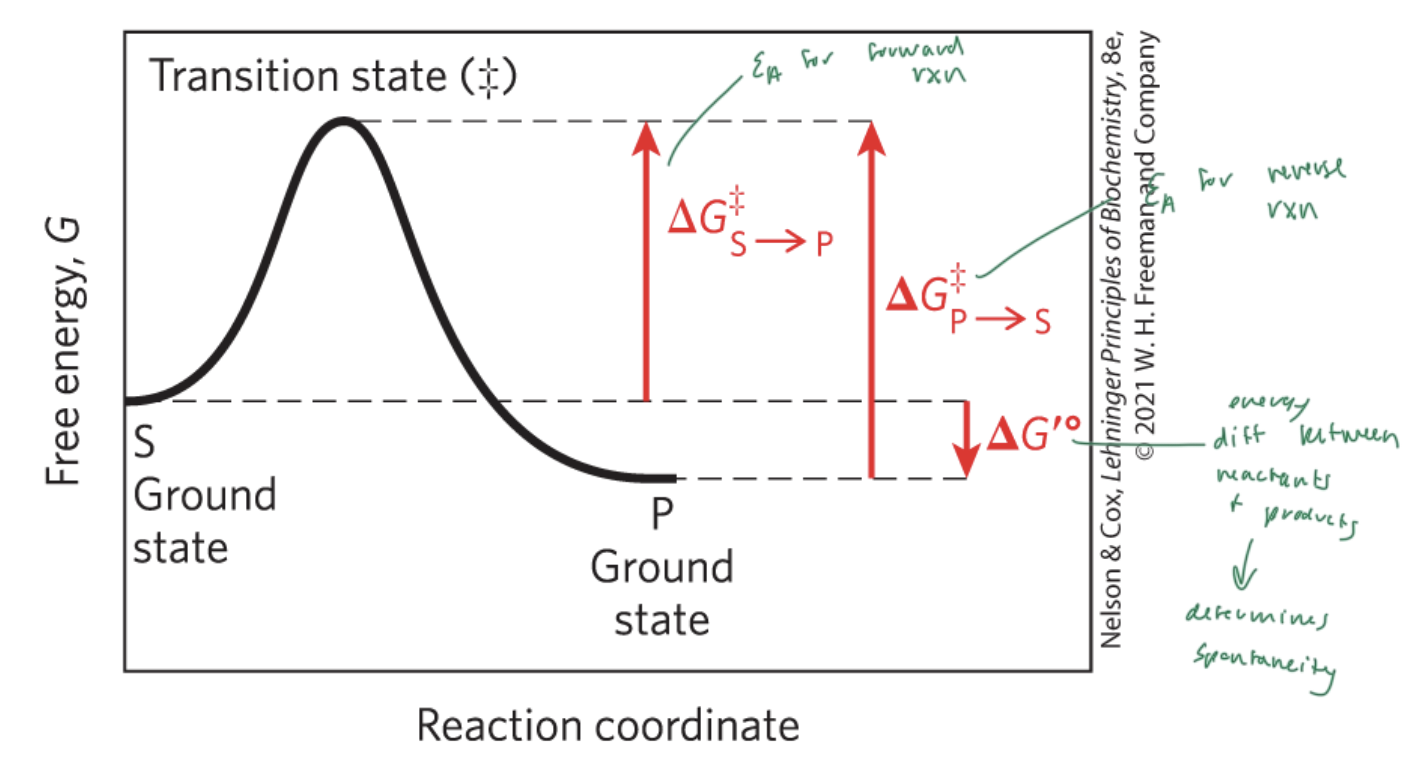
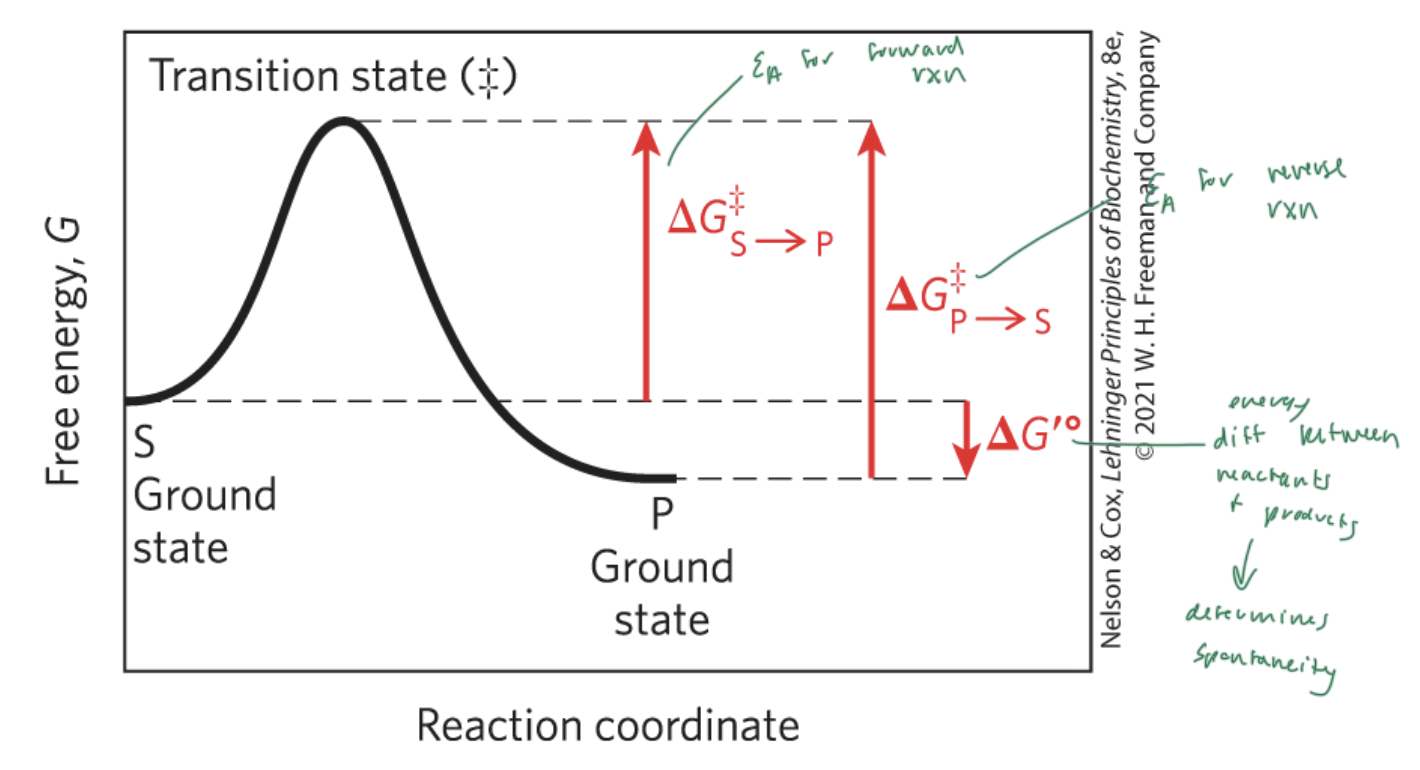
activation energy (Ea)
the energy required to start a rxn
the difference between energy levels of the ground state and transition state of a rxn
ΔG‡
lower activation energy means faster rxn
higher ΔG‡ means slower rxn
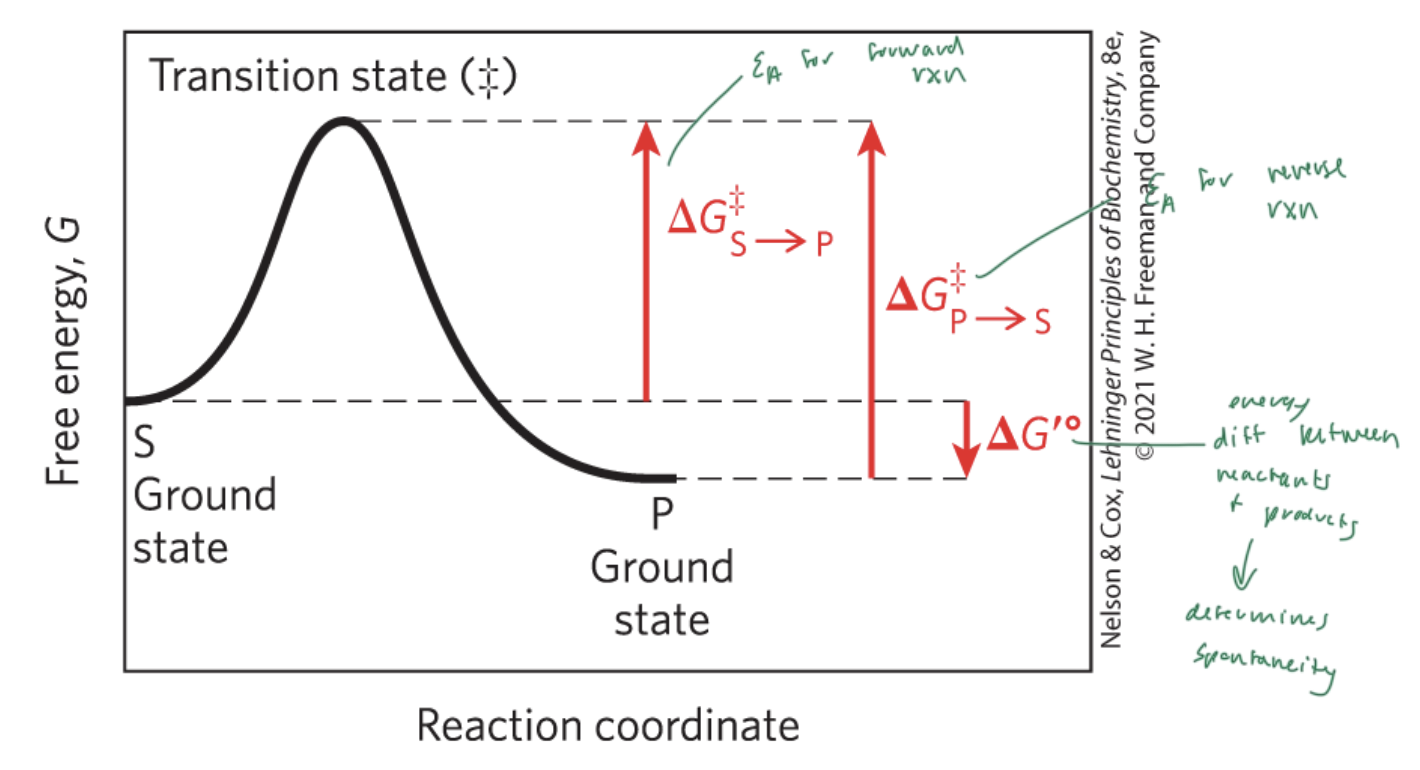
higher ΔG‡ means
slower rxn
ΔG‡S—>P
the Ea for the forward rxn
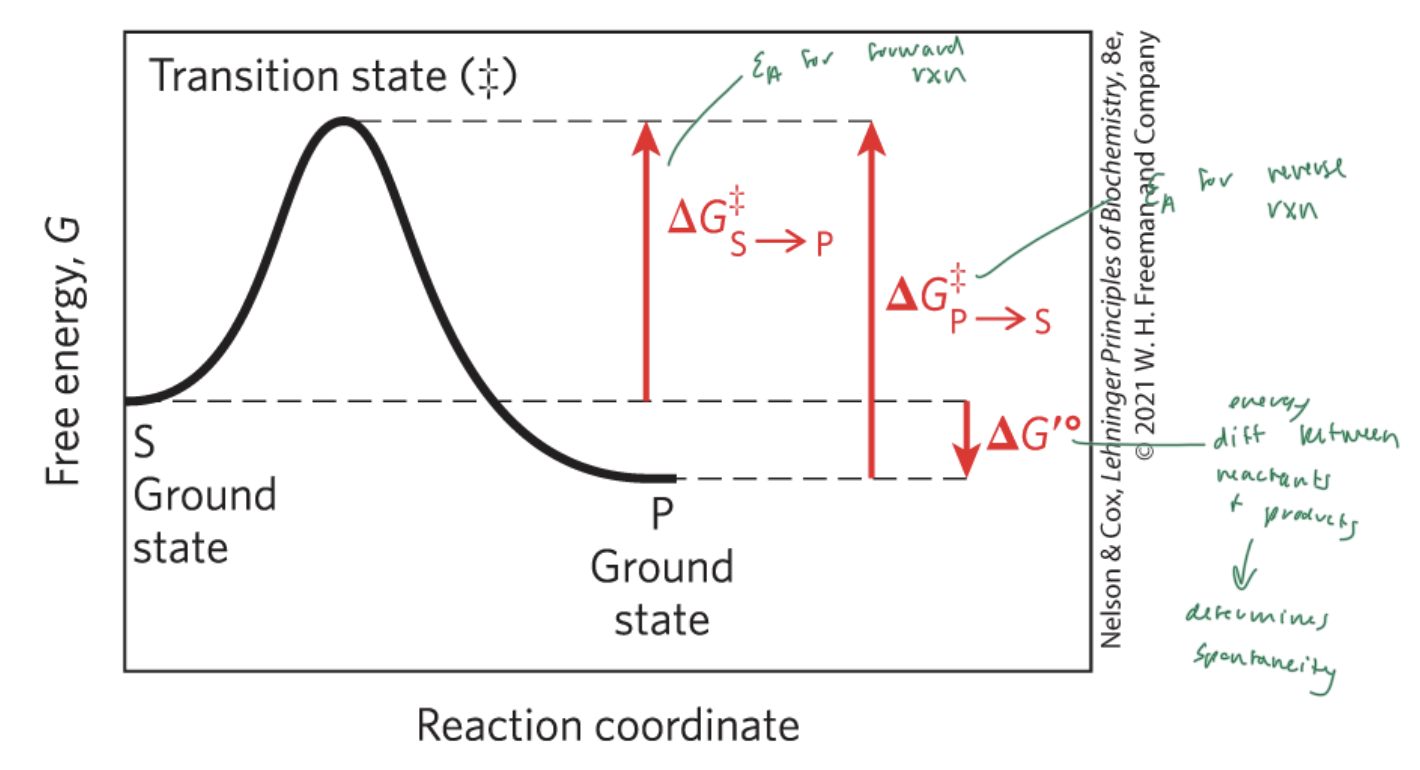
ΔG‡P—>S
the Ea for the reverse rxn
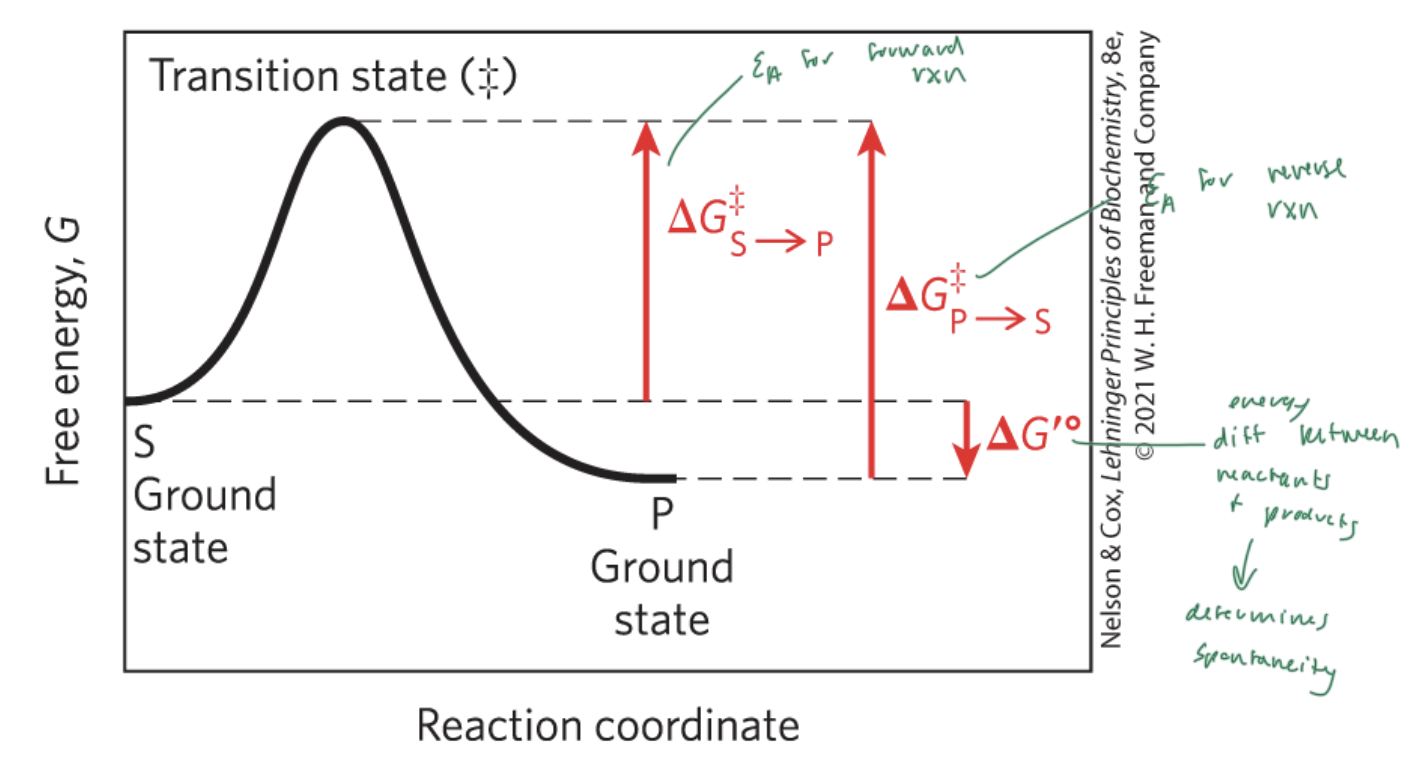
ΔG‡°
the energy difference between reactant and products
determines if a rxn is spontaneous
free energy ΔG
the amount of energy available to do work
in spontaneous reactions, _____ have less free energy than ____
products have less free energy than the reactants (so the forward rxn is favored)
ΔG will be neg (exergonic rxn)
even if ΔG° is neg, a rxn can be slow if
there is a lot of Ea
‡
transition state
positive ΔG means
the rxn is not spontaneous
backwards rxn is favored
endergonic rxn
if ΔG=0,
the reverse and forward rxn are equally favored
ΔG is the energy difference between
the substrate and the product
enzymes lower the
Ea and therefore the energy difference between the substrate and the transition state
NOT ΔG (which is the energy diff between the substrate and the product)
more Ea means
slower rxn
catalysts reduce
the Ea of a rxn
ΔG‡
free energy of activation
the energy required to convert 1 mol of substrate from the ground state to the transition state (‡)
the more molecules reaching the transition state means
the more likely product forms and therefore the faster the rxn reaches equilib
what binds better to the enzyme?
the transition state (of the substrate)
otherwise the rxn would not occur
it enables the product to actually be made since the substrate changes into the transition state so that it can bind to the enzyme better
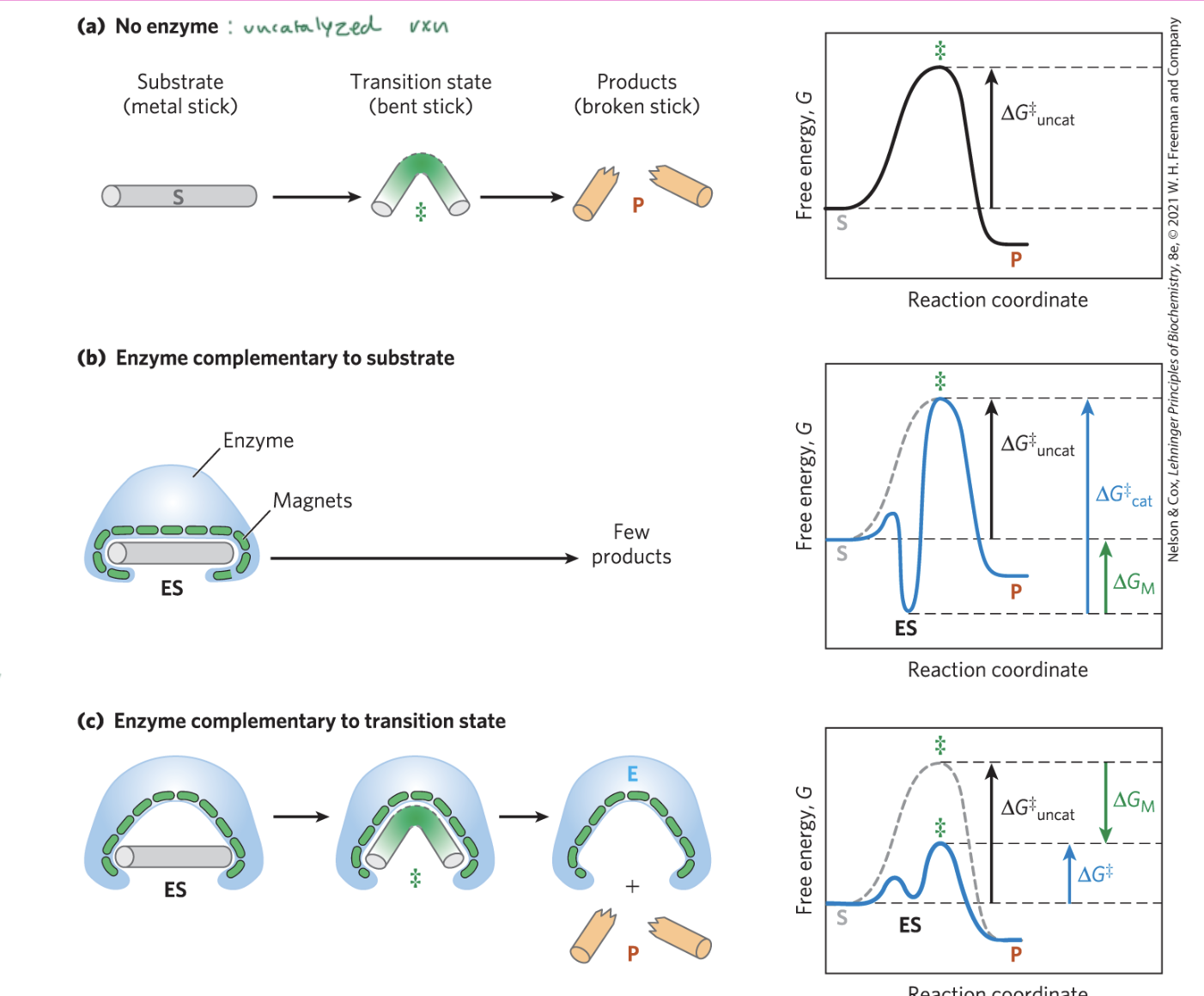
ways that enzymes are catalytic
proximity effect: bring substrates and active sites together
orientation effect: hold substrates at the exact distance and orientation necessary for rxn
catalytic effect: provide basic/acidic/etc side chain groups required for catalysis
energy effect: lower the energy barrier by inducing strain in bonds in the substrate molecule
how do enzyme-catalyzed rxns begin?
with the migration of the substrate into the active site to form an enzyme-substrate complex
mediated by weak noncovalent interactions and shape
induced fit can cause both the enzyme and the substrate to change shape
water can be displaced by the binding of the enzyme to the substrate
catalytic effect mechanisms of enzymes
acid/base catalysis: by removing/adding a proton to/from something to lower the free energy of the transition state
covalent catalysis: when the enzyme covalently binds to the substrate for a brief time (transiently) to initiate the formation of the enzyme-substrate interaction
metal ion catalysis: metal ions participate in the rxn mechanism
many enzymes use a combination of these methods
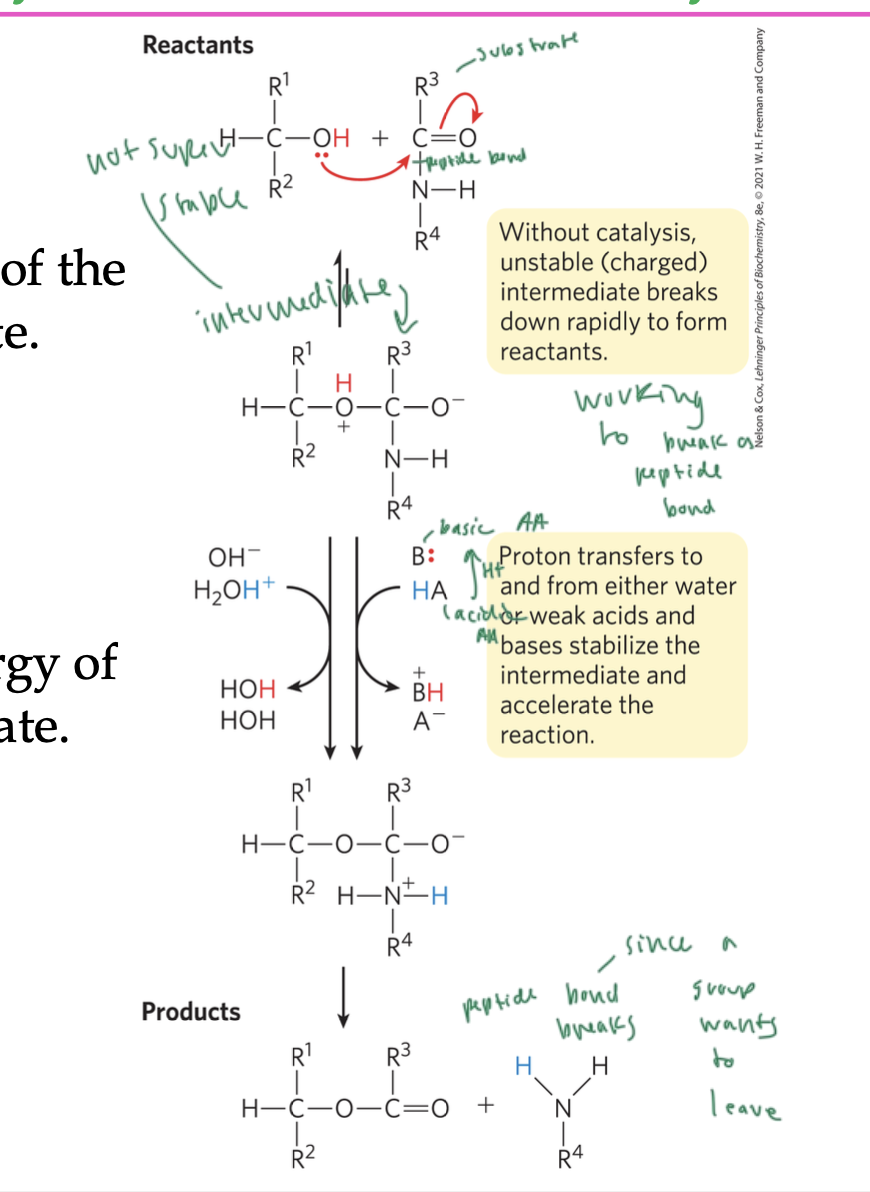
acid base catalysis
a proton transferring from an acid lowers the free energy of the rxn’s transition state (general acid catalysis)
a proton being removed by a base lowers the free energy of the rxn’s transition state (general base catalysis)
these rxns occur to stabilize intermediates to promote the continuation of a rxn
see image on slide
the substrate has a peptide bond
the intermediate has a charge (since when the enzyme and substrate combined, it was due to a proton transfer)
the proton transfers to and from either water or weak acids/base side chains to stabilize the intermediate, but this causes the peptide bond to be unstable
the product forms when the peptide bond is finally broken due to a leaving group leaving
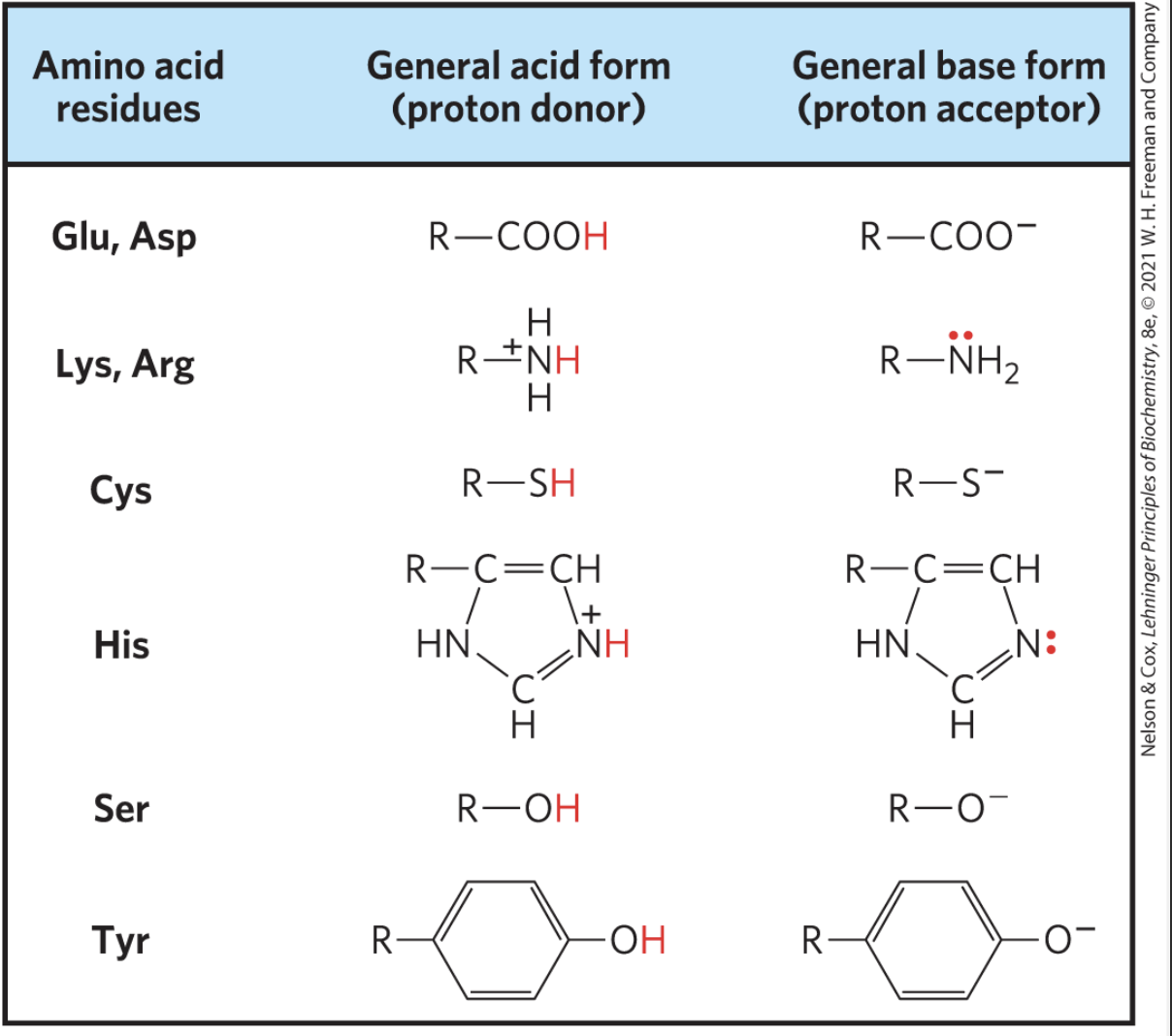
amino acids that serve as proton donors/acceptors based on protonation
Glutamic acid
aspartic acid
lysine
cysteine
histidine
serine
tyrosine
proton donors are in protonated form (have extra H, but aren’t always +)
how enzymes serve as covalent catalysts
a transient (temporary) covalent bond forms between the enzyme and the substrate
requires a nucleophile on the enzyme
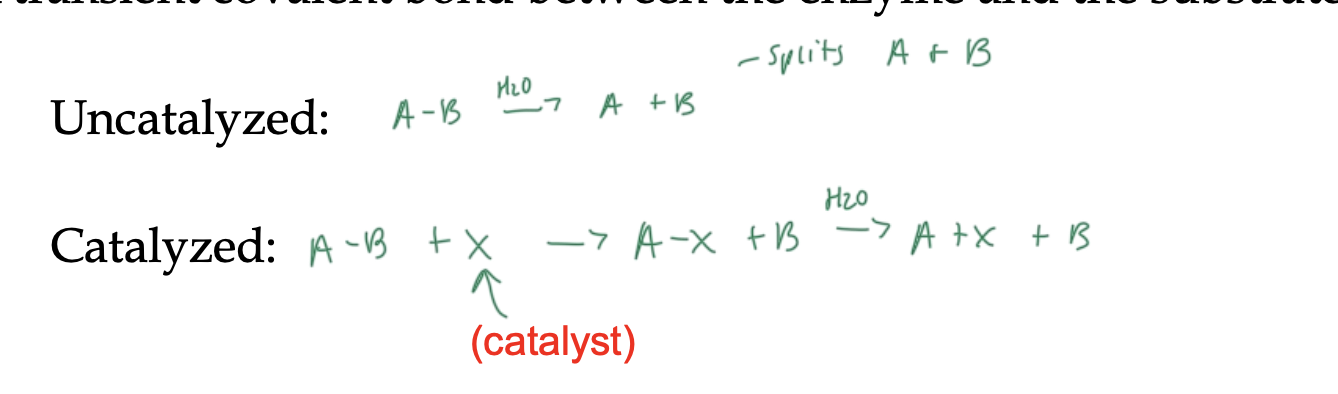
nucleophiles that can be on an enzyme for a covalent catalysis
reactive serine
thiol
amine
carboxyl
metal ion catalysis
when there is a coenzyme-like metal ion bound to the enzyme, which interacts with the substrate to facilitate binding of the substrate to the enzyme
metal ions help:
orient substrate for rxn
stabilize neg charges (since they’re often pos)
mediate redox reactions (keep other ions in certain charges)
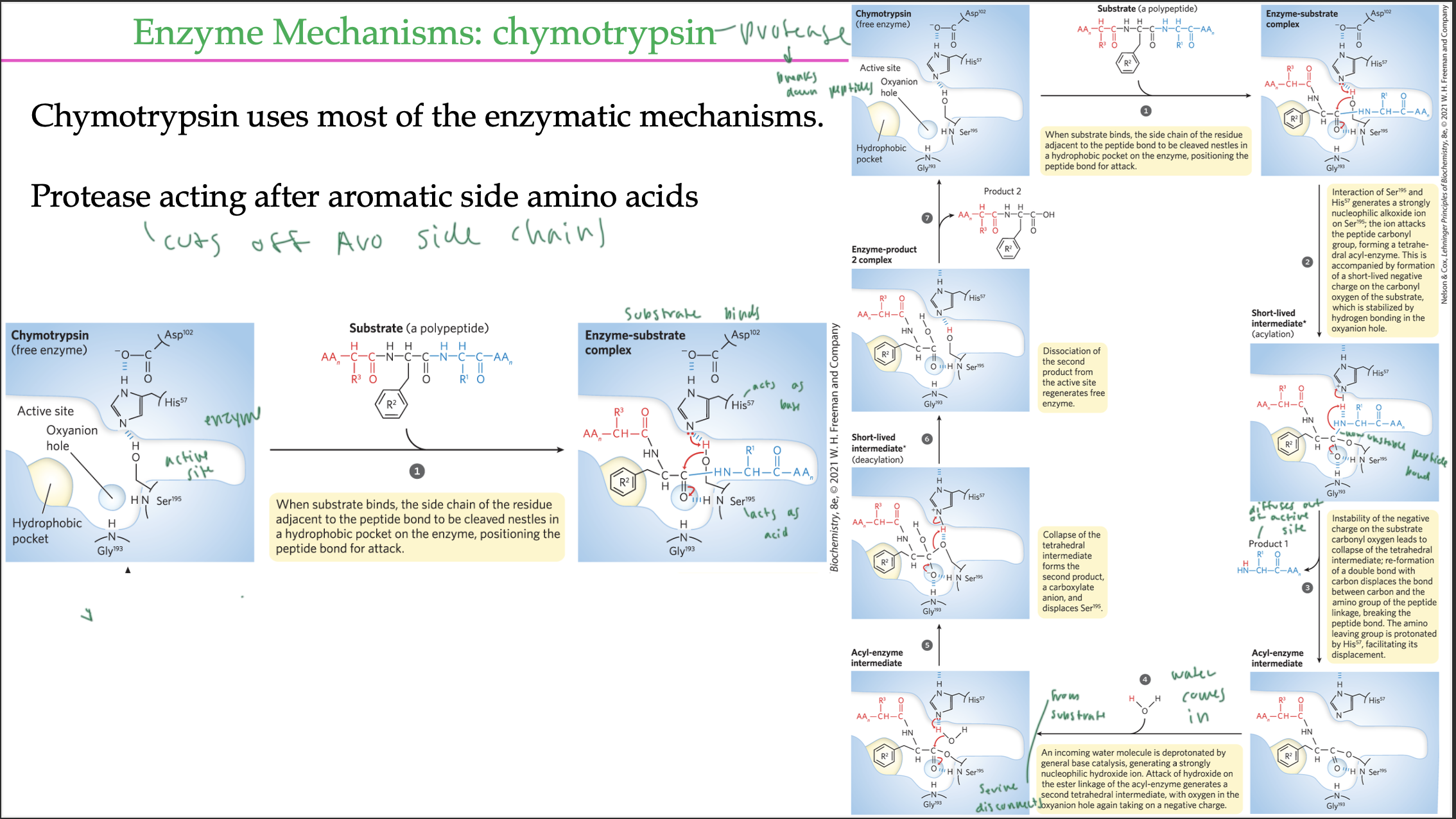
chymotrypsin is a
protease
chymotrypsin rxn pathway in the GI tract (ex of acid base covalent catalysis)
the protease chymotrypsin cuts off aromatic side chains in a peptide
His acts as base and Ser acts as the acid
peptide bond between C=O and HN (blue line)
N from His grabs the H from Ser’s OH (so His acts as base)
O of Ser attacks C of the C=O by the peptide bond of the substrate’s side chain, covalently attaching to it
now C-N bond (peptide bond) is destabilized, so N takes the H from His, so now N of the substrate broke the peptide bond
now part of the substrate is not held into active site, and it moves out
Ser is still covalently attached to the substrate in the active site of the enzyme
water comes in and breaks the Ser-substrate covalent bond so the substrate can leave
active site gets regenerated
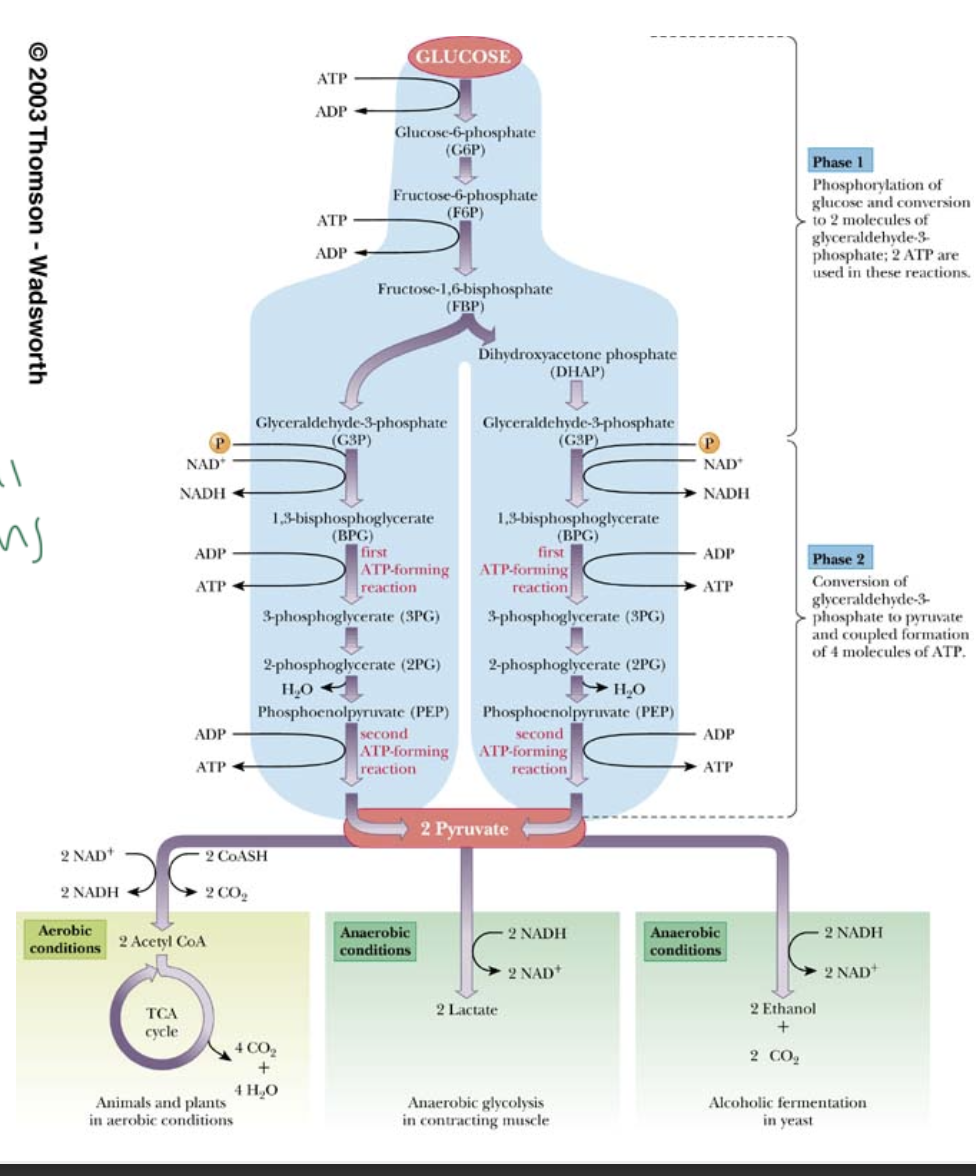
exergonic rxns are essential because
you need to counteract entropy
since cells cannot tolerate rxns that release huge amounts of energy/heat is released, what does it do instead?
many smaller rxns in a series of steps
ex: in glycolysis
types of enzymes
oxidoreductases: transfer electrons using H atoms of hydride ions
transferases: transfer the groups between two molecules
hydrolases: do hydrolysis (transfer functional groups to water)
lyases: cleave C-C, C-O, C-N or other bonds by elimination, leaving double bonds or rings, or add groups to double bonds (addition)
isomerases: transfer groups between molecules to yield isomeric forms
ligases: form C-C, C-S, C-O, and C-N bonds by condensation rxns coupled to cleave ATP or a similar cofactor
translocases: move molecules/ions across a membrane
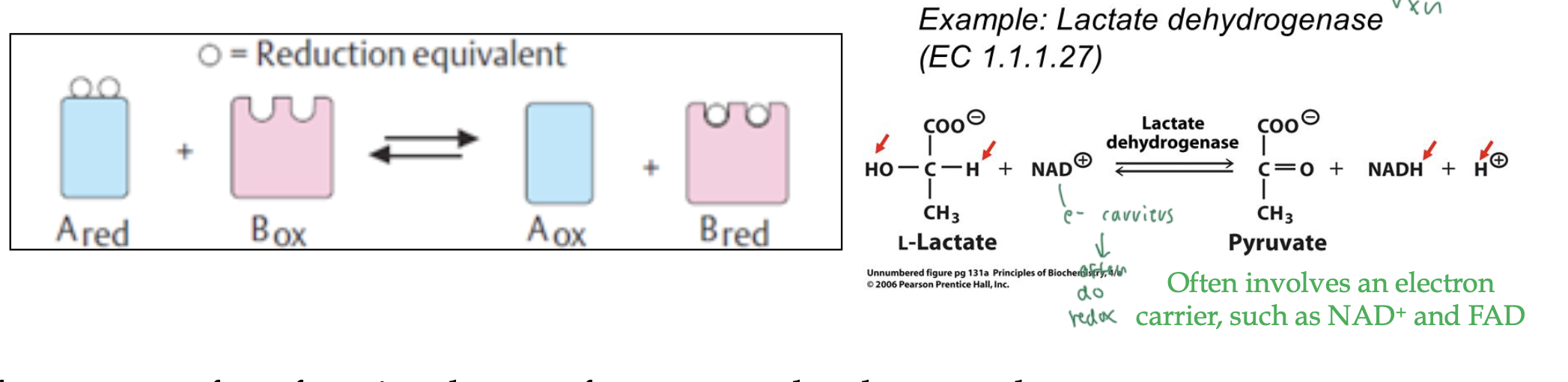
oxidoreductases
catalyze e- transfer in redox rxns
usually uses energy carriers like NAD+ to do redox rxns
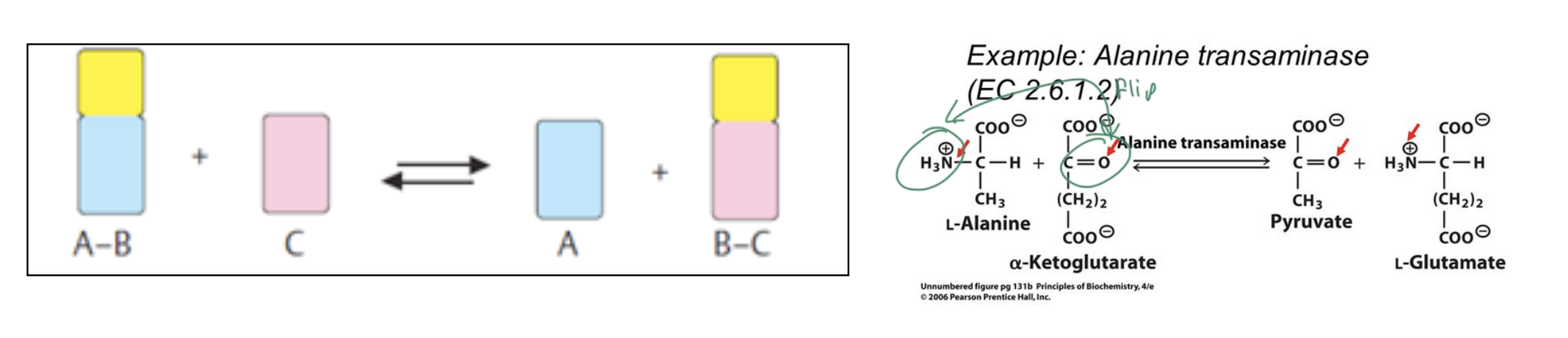
transferases
transfer a functional group from one molecule to another so that they switch groups
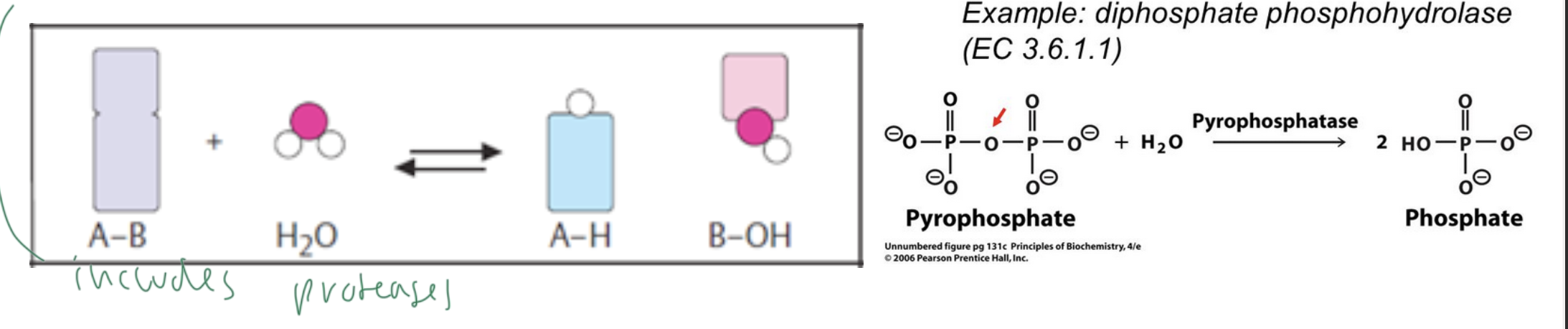
hydrolases
cause the cleavage if bonds using water (to break them apart)
aka do hydrolysis
includes proteases
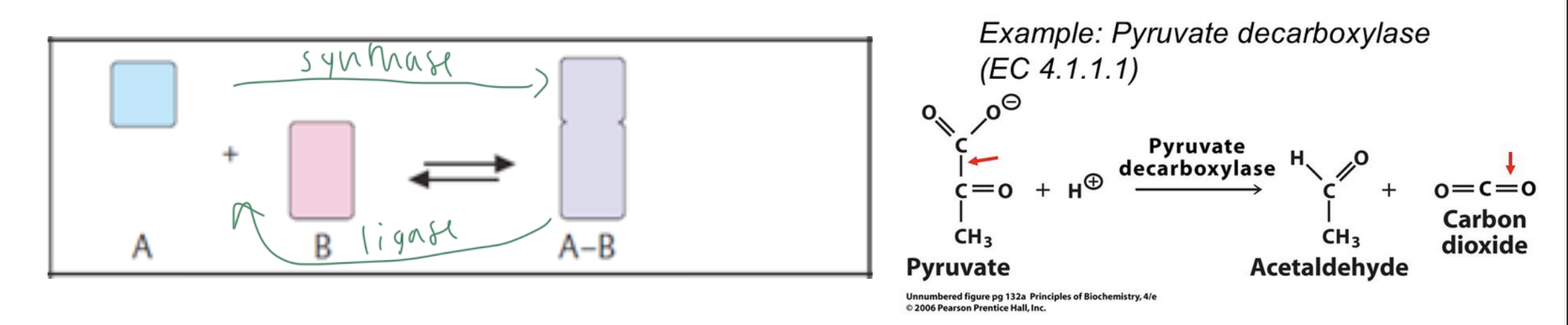
lyases/synthases
cleave or form a bond between two molecules (without using water or doing redox)
synthases do the forward rxn
liyases so the reverse rxn
isomerases
move groups within a singular molecule
intramolecular rearrangement to make a new molecule

ligases/synthetases
join two substrates together by cleaving ATP or another energy molecule
uses nucleotides for energy

translocases
move molecules or ions across membranes

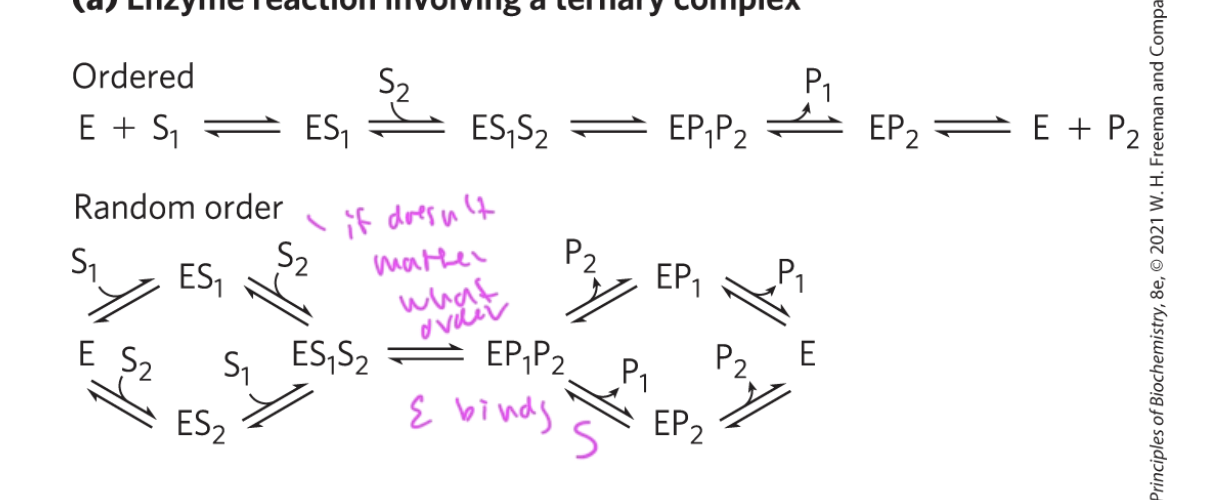
mechanism for enzymes that involve a ternary complex
can be ordered or be random
ordered: E must bind S1, THEN S2 before forming products (P1 and P2)
random: E can bing S1 and S2 in any order, but once both are bound to E, the products (P1 and P2) are formed in some manner
mechanism for enzymes that do NOT involve a ternary complex
the order matters
E binds S1 and forms P1 then binds S2 and makes P2
E gets changed after it makes P1, causing it to now bind S2
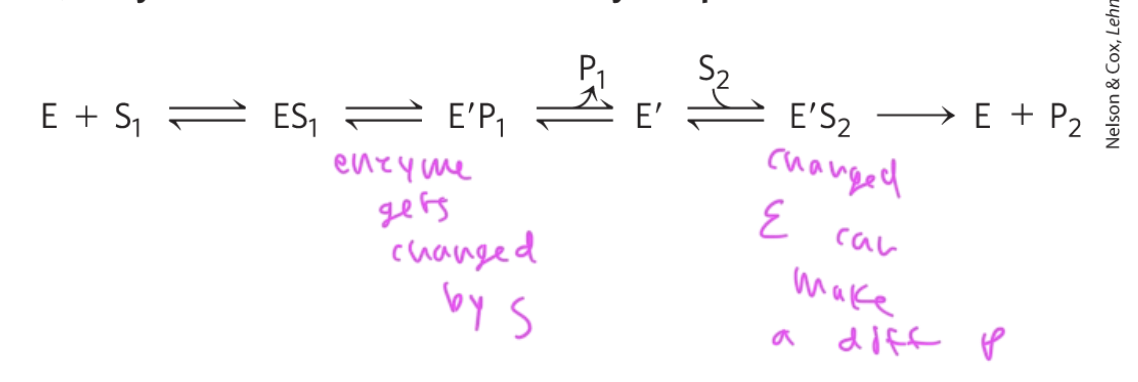
kinetics
the study of the rate at which compounds react
developed by Michaelis and Manten
velocity in kinetics is
the speed of the rxn
at low [S], v incr almost linearly with
an incr in [S]
but when [S] is already high, v increases by smaller and smaller amounts in response to an incr in [S]
vmax
the point at which increasing [S] does not increase velocity any further, so the graph plateaus
![<p>the point at which increasing [S] does not increase velocity any further, so the graph plateaus</p>](https://knowt-user-attachments.s3.amazonaws.com/5afabec4-ec5e-4041-98bf-58077ded2c8f.png)
rxn rate depends on
substrate conc
units ion velocity for enzymatic rxns
molarity/time
[Product]/seconds or minutes
velocity incr as [S] increases in a ____ manner
hyperbolic
can you use the vmax of two enzymes to compare them?
NO
vmax is only proportional to the conc of enzyme, so cannot be used to compare across different enzymes
turnover number (kcat)
the number of substrate molecules converted to product per enzyme per unit of time
only when E is SATURATED with substrate
kcat= Vmax/[E]
has units of /s or /min
Vmax=
[E] (Kcat)
two enzymes catalyzing different reactions could have the same
kcat
but their rates of their uncatalyzed rxns are likely different
so their rate enhancements likely vary greatly
Michaelis-Manten Constant
Km= [E][S]/[ES]
km is a
constant
since once the rxn starts, [ES] is constant
[ES] can break down and do reverse rxn too
steady-state assumption
idea that the rate of formation of ES is equal to its breakdown
so [ES] is constant
![<p>idea that the rate of formation of ES is equal to its breakdown</p><ul><li><p>so [ES] is constant</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/8f149d33-4bb9-415e-8734-9e5b101a4454.png)
enzymes can have diff ____ for diff ___
Kms for diff substrates
steady state assumption equation
k1[E][S]= k-1[ES]+K2[ES]
basically Km=[E][S]/[ES]
a small Km indicates
tighter binding
since less substrate is unbound
Km= [E][S]/[ES]
Km is
the [S] at which half of the enzyme molecules have substrate bound in their active sites to produce ES
so Km=[S] since [E]=[ES]
Km is inversely proportional to
how well an enzyme binds its substrate
does km equal ½ Vmax?
NO
Km is the [S] at ½ Vmax
![<p>NO</p><ul><li><p>Km is the [S] at ½ Vmax</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/db4c21e4-1a68-4650-8a61-2a388d0f22ee.png)
Michaelis-Manten equation expressed in terms of Vmax and Km
Vo= [(Vmax)[S]]/Km + [S]
this equation graphs as a hyperbola
![<p>Vo= [(Vmax)[S]]/Km + [S]</p><ul><li><p>this equation graphs as a hyperbola</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/5a78639d-6c58-453d-8eb4-ef965ab007ff.png)
Vmax is the velocity of the rxn when
substrate is infinitely available
so [S]>Km
when [S]=Km, vmax is
at ½ vmax

catalytic efficiency
kcat/Km
since affinity is inversely proportional to Km and kcat reflects rate of formation
a perfect enzyme would have
a high kcat/Km ratio
since you want Km low and kcat high
few enzymes are “perfect”
why is enzyme inhibition helpful?
for regulation
don’t want enzymes active all the time
many drugs and toxic agents act to
inhibit enzymes
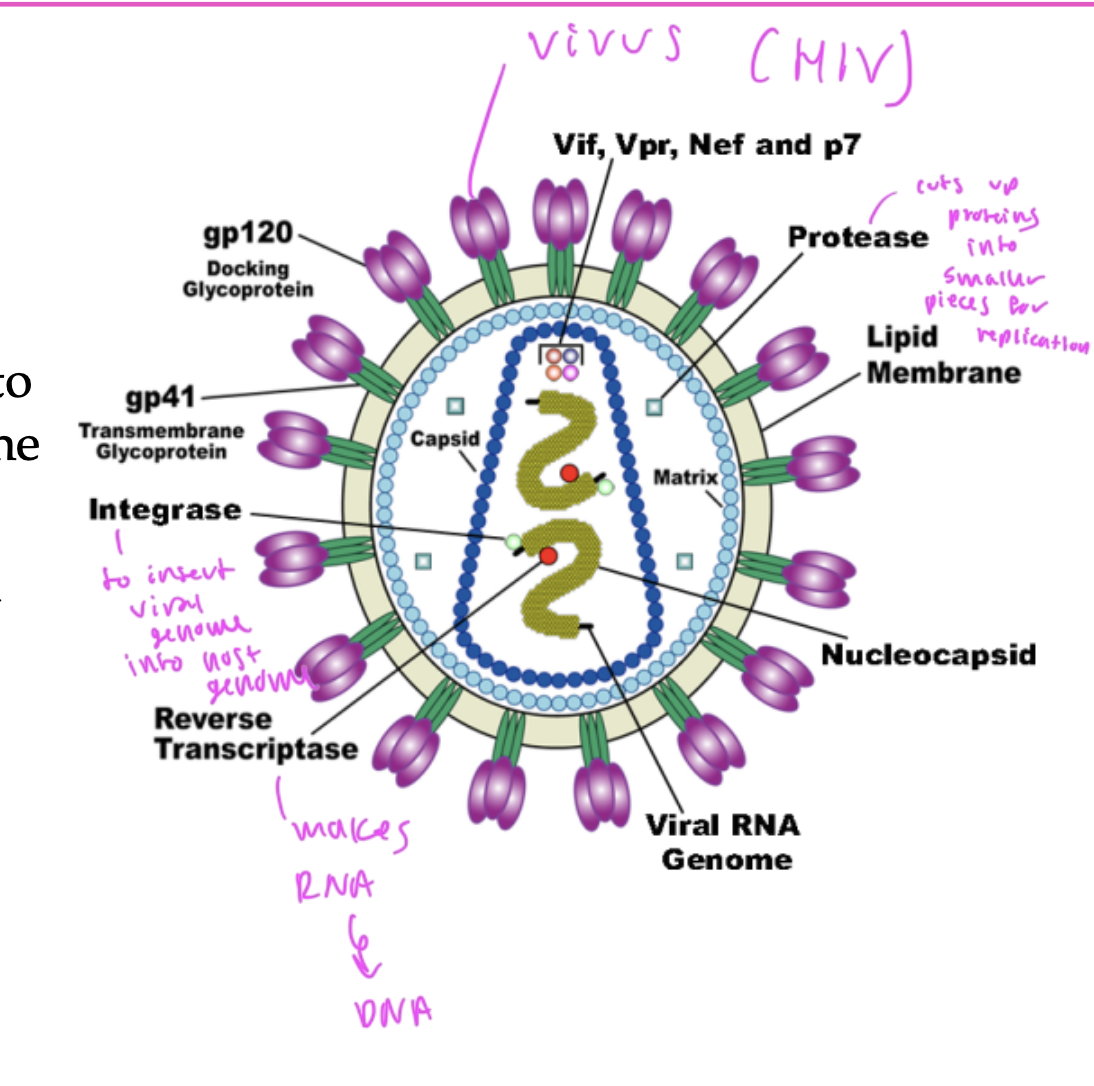
AIDS treatment
scientists are trying to develop specific inhibitors to block the enzymes that are unique to HIV
integrase: inserts the viral genome into the host genome
protease: cuts up proteins into smaller pieces to be replicated
reverse transcriptase: turns RNA into DNA
these are the 3 enzymes used by the HIV virus that you want to inhibit
enzyme inhibitors decr
the activity of the enzyme
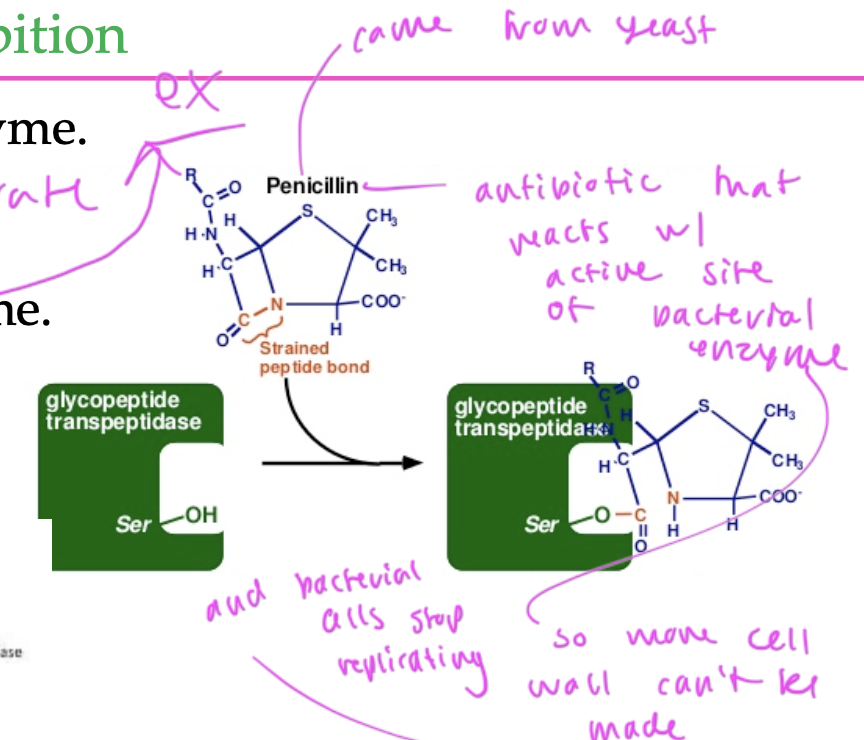
irreversible inhibitors (inactivators)
covalently attach to the enzyme so that substrate cannot bind
often are powerful toxins
ex: penicillin is an antibiotic that reacts with the active site of a bacterial enzyme to inhibit it so no more bacterial cell wall can be made and the bacterial cells stop replicating
reversible inhibitors
bind to the enzyme reversibly (so can easily dissociate, therefore don’t covalently bond but use molecular complementarity stuff)
are often structural analogs of substrates or products that they are replacing/inhibiting
can bind to the free enzyme to prevent it from binding to the substrate OR bind to the ES complex to prevent it from creating the final product
used as drugs to slow down an enzyme
2 types of reversible inhibitors
competitive and not competitive
competitive: when the inhibitor beats out the substrate to bind to the enzyme at the active site
Not competitive: when the inhibitor interacts with another spot of the enzyme (not the active site) OR does both
what types of inhibition affect the Km?
Competitive, uncompetitive, and mixed types of enzyme inhibition affect the apparent Km (Michaelis constant), while non-competitive inhibition typically does not
competitive: incr Km by decr substrate binding affinity
uncompetitve: decr Km by incr the enzyme’s affinity for the substrate
mixed: can either increase or decrease apparent Km, depending on whether the inhibitor preferentially binds to the free enzyme or the enzyme-substrate complex
non-competitive: binds equally to the free enzyme and the enzyme-substrate complex, causing the Km to remain unchanged
competitive inhibition
when the inhibitor binds in place of the substrate in the active site, reducing the productivity of the enzyme
the inhibitor does not affect the catalysis of substrate forming product
if [S]>[I], enzyme will bind more preferentially to the substrate than the inhibitor
![<p>when the inhibitor binds in place of the substrate in the active site, reducing the productivity of the enzyme</p><ul><li><p>the inhibitor does not affect the catalysis of substrate forming product</p></li><li><p>if [S]>[I], enzyme will bind more preferentially to the substrate than the inhibitor</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/58c83933-4ca7-435f-b835-32a41aff51e8.png)
see how substrate conc affects competitive inhibition
the inhibitor acts as a transition state analog
if [S]=[I], then the inhibitor or substrate bind 50/50
when more substrate gets added, it binds more preferentially than the inhibitor
![<p>the inhibitor acts as a transition state analog</p><p>if [S]=[I], then the inhibitor or substrate bind 50/50</p><p>when more substrate gets added, it binds more preferentially than the inhibitor</p>](https://knowt-user-attachments.s3.amazonaws.com/40e37cfd-51d9-40f2-aede-2b9f87847763.png)
methotrexate
a competitive inhibitor and chemo drug
competes with dihydrofolate (the substrate) to inhibit nucleotide synthesis
basically outcompetes the dihydrofolate so that new nucleotides cannot be made by the enzyme and slows down cell division for a cancer pt to defeat the tumor
but also leads to side effects since your other cells can’t divide either, makes you feel ill
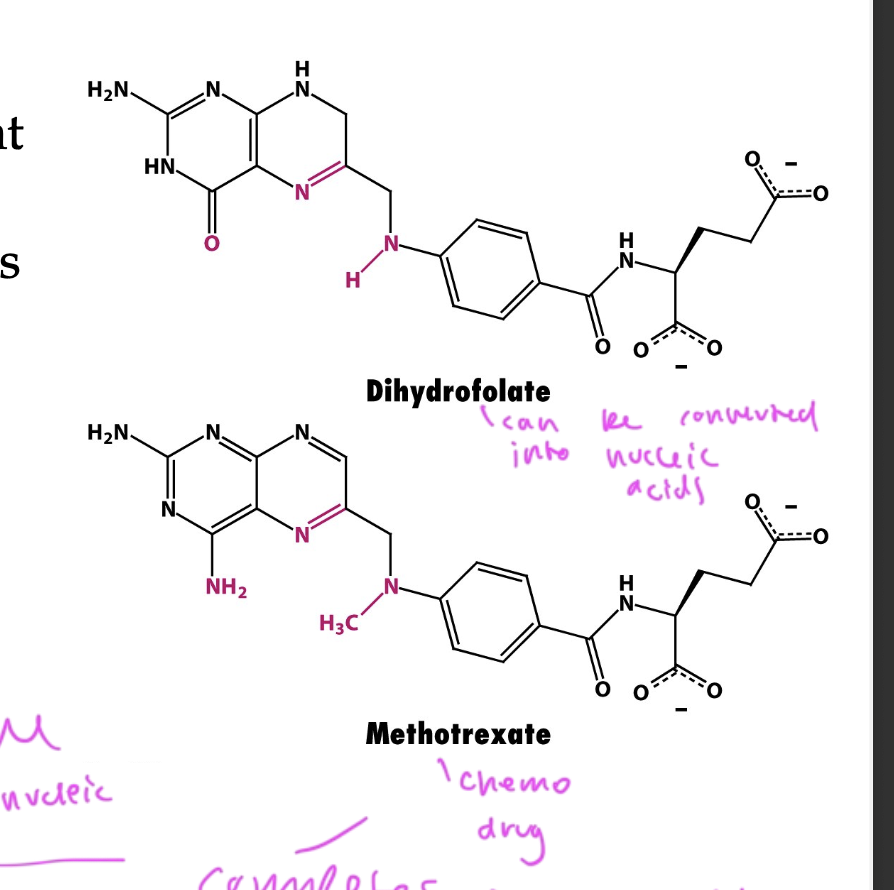
dihydrofolate
can be converted into nucleic acids for replication
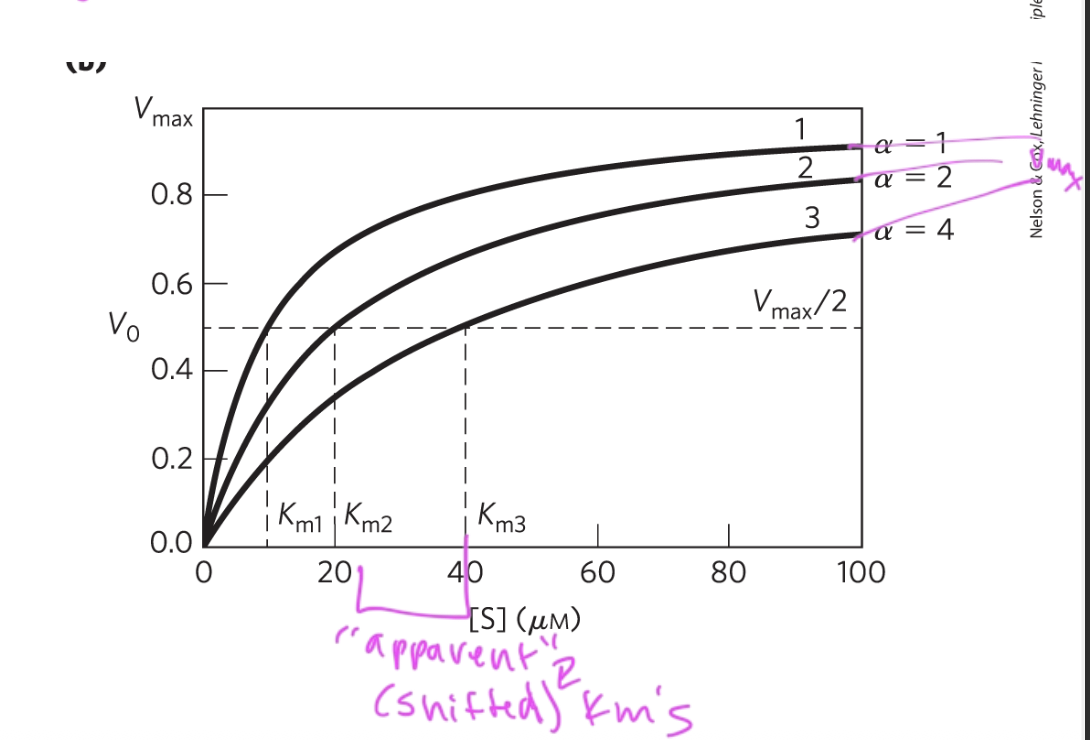
competitive inhibitors’ affect on Km
incr Km, since the inhibitor decr enzyme affinity for substrate (so more substrate is needed to obtain the same rxn rate)
but all values still reach the same vmax, which is unchanged
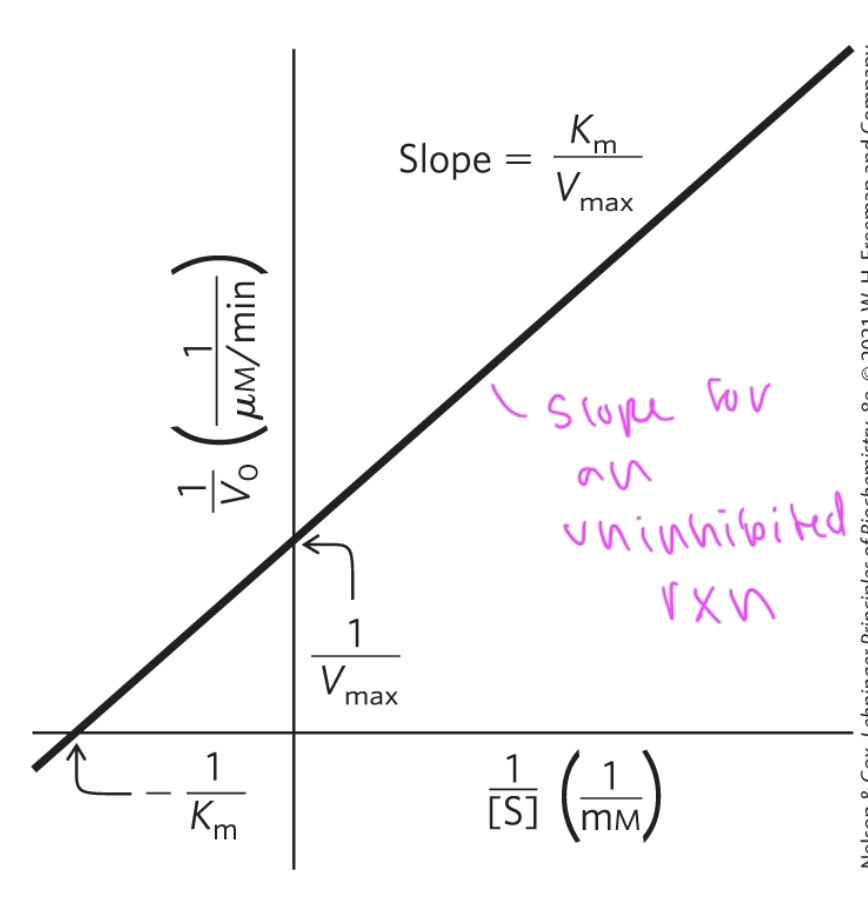
Lineweaker-Burk plots
can be used to distinguish between diff types of inhibition
the slope of the line shows Km/Vmax (which is the inverse of the Michaelis-Manten equation)
the y-intercept is 1/Vmax
x intercept is -1/Km
plot shows unhibited rxn
Lineweaver-Burk plot for competitive inhibited rxn
slope (Km/Vmax) incr as [I] incr
intercept on x-axis (-1/Km) changes to show an increased Km when the inhibitor is present
the Vmax remains unchanged
slope incr as [I] incr
![<ul><li><p>slope (Km/Vmax) incr as [I] incr</p></li><li><p>intercept on x-axis (-1/Km) changes to show an increased Km when the inhibitor is present</p></li><li><p>the Vmax remains unchanged</p></li><li><p>slope incr as [I] incr</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/71b1ec4d-58d6-4415-b1f9-305c2cf1a7c7.png)
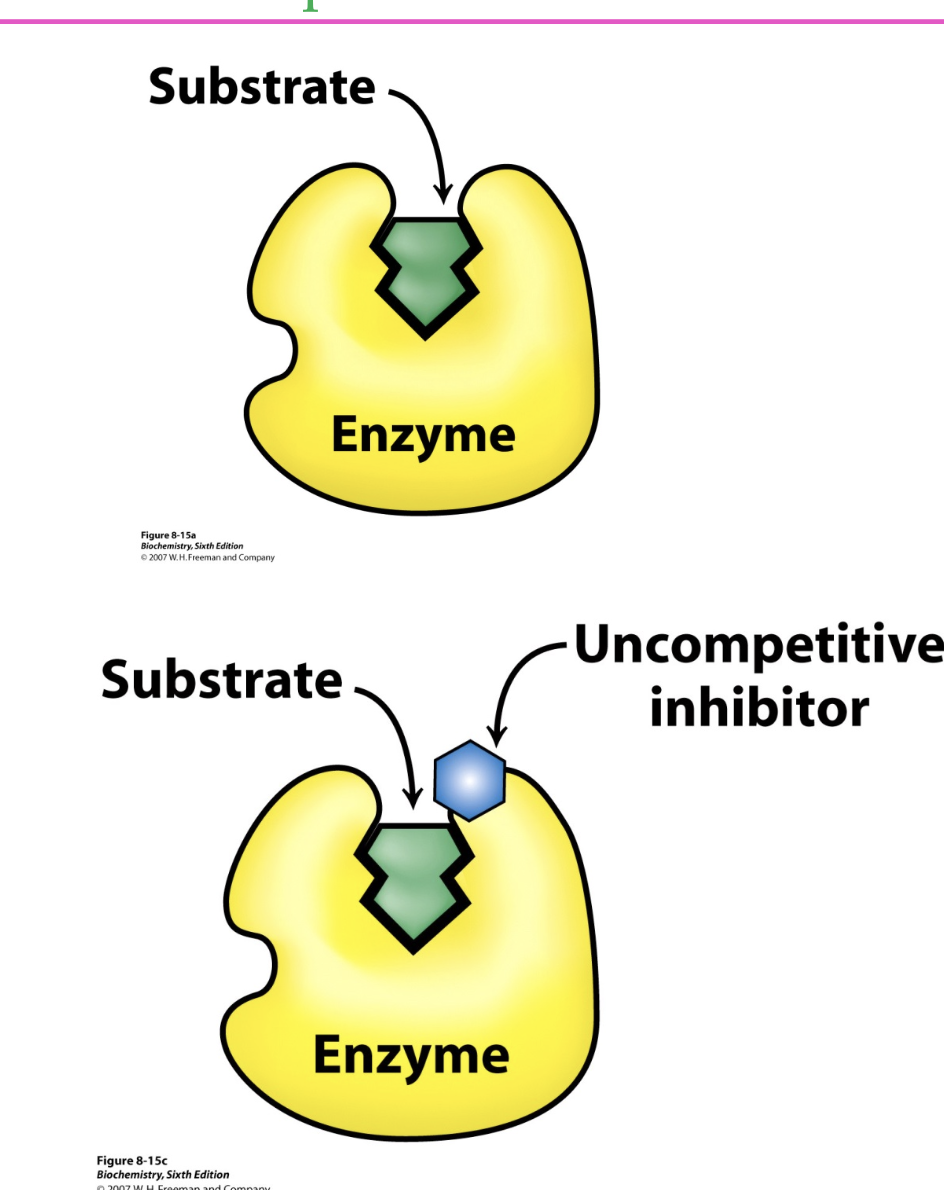
uncompetitive inhibition
the inhibitor binds to the whole ES complex, to stop it from forming product
the ESI complex can NOT form products
the binding site for the inhibitor isn’t created until the enzyme binds to the substrate
inhibition can NOT be overcome by adding more S