Genetics Cumulative Final - Hughes
1/328
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
329 Terms
Where does the eukaryotic RNA polymerase II bind?
a. core promoter
b. regulatory promoter
c. enhancer
d. intron
a. core promoter
Where does pre-mRNA splicing occur?
a. in the mitochondria
b. in the cytoplasm
c. on the ribosome
d. in the nucleus
d. in the nucleus
20 amino acids are encoded by 61 sense codons, this means what?
a. There are isoaccepting tRNAs
b. There is wobble
c. The code is degenerate
d. All of the above.
c. The code is degenerate
What ensures that any particular tRNA only ever carries one particular amino acid?
a. the ribosome.
b. the aminoacyl-tRNA synthetase
c. the genetic code
d. isoaccepting tRNAs
b. the aminoacyl-tRNA synthetase
During the elongation phase of translation, what part of the ribosome receives the next aminoacyl-tRNA ?
a. A site
b. E site
c. P site
d. codon
e. anti-codon
a. A site
Where does the small subunit of the ribosome first bind to an E. coli mRNA?
a. AUG start codons
b. -10 and -35 consensus sequences
c. Shine Dalgarno sequences
d. 5' caps
c. Shine Dalgarno sequences
What is the structure of the DNA binding domain of the lac I protein?
a. leucine zipper
b. zinc finger
c. helix-turn-helix
d. acidic activation
c. helix-turn-helix
What is true of the genes that are part of a single bacterial operon?
a. Encode proteins with the same function
b. Encode repressors and aporepressors
c. Translated into a single enzyme
d. Transcribed from a single promoter
d. Transcribed from a single promoter
Where does the lac I protein bind?
a. lac P
b. lac O
c. lac Z
d. lac Y
b. lac O
What causes induction of the lac operon?
a. lac I binding to cAMP
b. cAMP binding to CAP
c. allolactose binding to CAP
d. allolactose binding to lac I
d. allolactose binding to lac I
EMS adds an ethyl group to Guanine, and ethylguanine pairs with thymine. What kind of mutation will result if this damage is not repaired before DNA replication?
a. GC to AT transversion
b. GC to AT transition
c. GC to TA transversion
d. GC to TA transition
b. GC to AT transition
How does the E. coli mismatch repair system distinguish between the template strand and newly-synthesized strand?
a. presence of uracil
b. apurinic sites
c. adenosine methylation
d. apyrimidinic sites
c. adenosine methylation
CRISPR/Cas9 can be a tool for changing DNA sequence when the double strand DNA cut is repaired how?
a. Mismatch repair
b. Homology-directed repair
c. Nonhomologous end joining
d. Base-excision repair.
b. Homology-directed repair
When a plasmid has been engineered for blue/white color selection of recombinant DNA, where is the DNA ligated into the plasmid?
a. In the lacZ gene
b. In the ori of replication
c. In the selectable marker
d. In the promoter.
a. In the lacZ gene
In fluorescent di-deoxy sequencing, where is the fluorescent dye found?
a. On the deoxyNTPs
b. On the DNA polymerase
c. On the dideoxyNTPs
d. On the primer.
c. On the dideoxyNTPs
To find the gene responsible for an inherited disorder, researchers would use what approach?
a. Plasmid cloning
b. DNA sequencing
c. PCR of microsatellites
d. SNP genotyping
d. SNP genotyping
Which of the following is one characteristic of the 5' end of a eukaryotic mRNA?
methylation
acetylation
adenosine residues
Shine Dalgarno sequence
methylation
(a methyl group is added on to the second carbon)
How is the poly-A tail of a eukaryotic mRNA added?
By a special enzyme
By the RNA polymerase
By splicing
By cleavage
By a special enzyme
(the 3' end is cleaved but then poly-A-polymerase adds a chain of adenine nucleotides to create the poly-A tail)
In what ways is a primary transcript modified to produce a mature tRNA?
All of these ways
Clevage of a long transcript
Chemical modification of bases
Enzymic addition of CCA
All of these ways
What is the most likely explanation for a suite of proteins that share the same amino acid sequence at the beginning (amino end) but vary in length?
Multiple 3' cleavage sites
Alternative splicing
Variable processivity
RNA polymerase failure
Multiple 3' cleavage sites
There are 61 sense codons that code for a total of 20 amino acids; because of this the code is called what?
Degenerate
Isoaccepting
Wobble
Determined
Degenerate
After peptidyl transferase activity, and translocation of the ribosome, what is the location of the tRNA attached to the growing peptide?
P site
A site
E site
P site
How do E. coli coordinate the expression of functionally-related genes?
Transcribed from same promoter
Shared response elements
Have related inducers
Using cAMP/CAP
Transcribed from same promoter
The lac operon is described as " inducible "; what does this mean?
It is usually off, but can be turned on.
It is usually on, but can be turned off.
It is under positive control.
It is under negative control.
It is usually off, but can be turned on.
What is the function of the Lac I protein that enables it to function as a repressor?
Binds to DNA
Binds to beta galactosidase
Binds to lactose
Binds to RNA polymerase
Binds to DNA
When the promoter of the lac operon is mutated so that it no longer works (a P minus mutation), how is this expected to change the expression of the operon?
It will be uninducible
It will be inducible
It will be constitutive
It will be repressible
It will be uninducible
Which of the following elements of the lac operon is trans operating?
Lac I
Lac P
Lac O
Lac Z
Lac I
What environmental conditions will maximize the rate of expression of the lac operon?
Low glucose, high lactose
Low glucose, low lactose
High glucose, high lactose
Low glucose, low lactose.
Low glucose, high lactose
Which of the following is a component of the basal transcription apparatus of eukaryotic structural genes?
All of them
RNA polymerase II
General transcription factors
Mediator of transcription
All of them
Which of the following modifications is likely to increase transcription of a eukaryotic gene?
Histone acetylation
DNA methylation
Chromatin condensation
Histone deacetylation
Histone acetylation
Where do transcriptional activating proteins bind to increase the rate of transcription in eukaryotes?
Enhancer sequences
Insulator sequences
Core promoter
RNA polymerase II
Enhancer sequences
Some human genetic diseases are caused by problems replicating repetitive regions within a gene, often CAGCAGCAG........CAG. Disease arises when these repeats
increase in number
decrease in number
undergo substitutions
undergo inversions
increase in number
Which of the following mutations is most likely to have very negative effects?
Insertion of 2bp in exon 1
Insertion of 2bp in intron 1
Inversion of 2bp in exon 1
Inversion of 2bp in intron 1
Insertion of 2bp in exon 1
When a substitution mutation changes a sense codon to a non-sense codon, what will be the result?
Premature end of translation
Premature end of transcription
Error in replication
Unlikely to have any effect
Premature end of translation
The Ames test for potential carcinogens requires that the Salmonella bacteria be incubated with what?
All of these
Histidine
Suspected mutagen
Liver extract
All of these
Bacteria produce their own endonuclease called restriction enzymes; why do bacteria produce these?
Restrict which bacteriophage can invade the cell
Cut the bacterial genome at specific sites
Require a matching set of methylases
Enable cloning into plasmids
Restrict which bacteriophage can invade the cell
How does the Southern blot (of electrophoretic gels) reveal the location of bands of the DNA of interest?
Probing with complementary DNA
Staining with ethidium bromide
Staining with antibodies
Location of blue dyes on gel
Probing with complementary DNA
At the end of an experiment where DNA has been cloned into a plasmid, and transformed into E. coli, they are plated out on media that contains Ampicillin, X-gal, and IPTG. What can you conclude about the blue colonies that grow?
They contain a non-recombinant plasmid
They do contain a recombinant plasmid
They are lac minus
They do not contain a plasmid
They contain a non-recombinant plasmid
In the classic Sanger-Harris, chain termination, method of sequencing, what is the special component that allows the technique to reveal the series of As Cs Gs and Ts?
dideoxynucleotide triphosphate
nucleotide triphosphate
single primer
thermostable DNA polymerase
dideoxynucleotide triphosphate
The polymerase chain reaction produces many copies of a very specific part of the template DNA; the length of those copies can be calculated as:
5' end of one primer to 5' end of other primer
3' end of forward primer to 5' end of reverse primer
3' end of one primer to 3' end of other primer
3' end of reverse primer to 5' end of forward primer
5' end of one primer to 5' end of other primer
The approach that the FBI uses to genotype samples from crime scenes and match them to suspects uses "micro satellite loci", these are ideal for this purpose because:
There are many polymorphic micro satellite loci
They are fully dominant
They are easily assessed by sequencing
All of these make them ideal
There are many polymorphic micro satellite loci
How does gene expression compare between bacteria and eukaryotes?
Bacteria often use negative control, eukaryotes often use positive control
Bacteria often use positive control, eukaryotes often use negative control
Both kingdoms largely use negative control
Both kingdoms largely use positive control.
bacteria often use negative control, eukaryotes often use positive control
Linkage analyses that try to find the locus, or loci, that cause human genetic disease rely on what source of data?
SNPs
Microsatellite loci
Blood type
SINEs
SNPs
One control over expression of the tryptophan operon is "attenuation", what is this?
Early termination of transcription
Binding of the repressor
Early termination of translation
Binding of tryptophan
Early termination of transcription
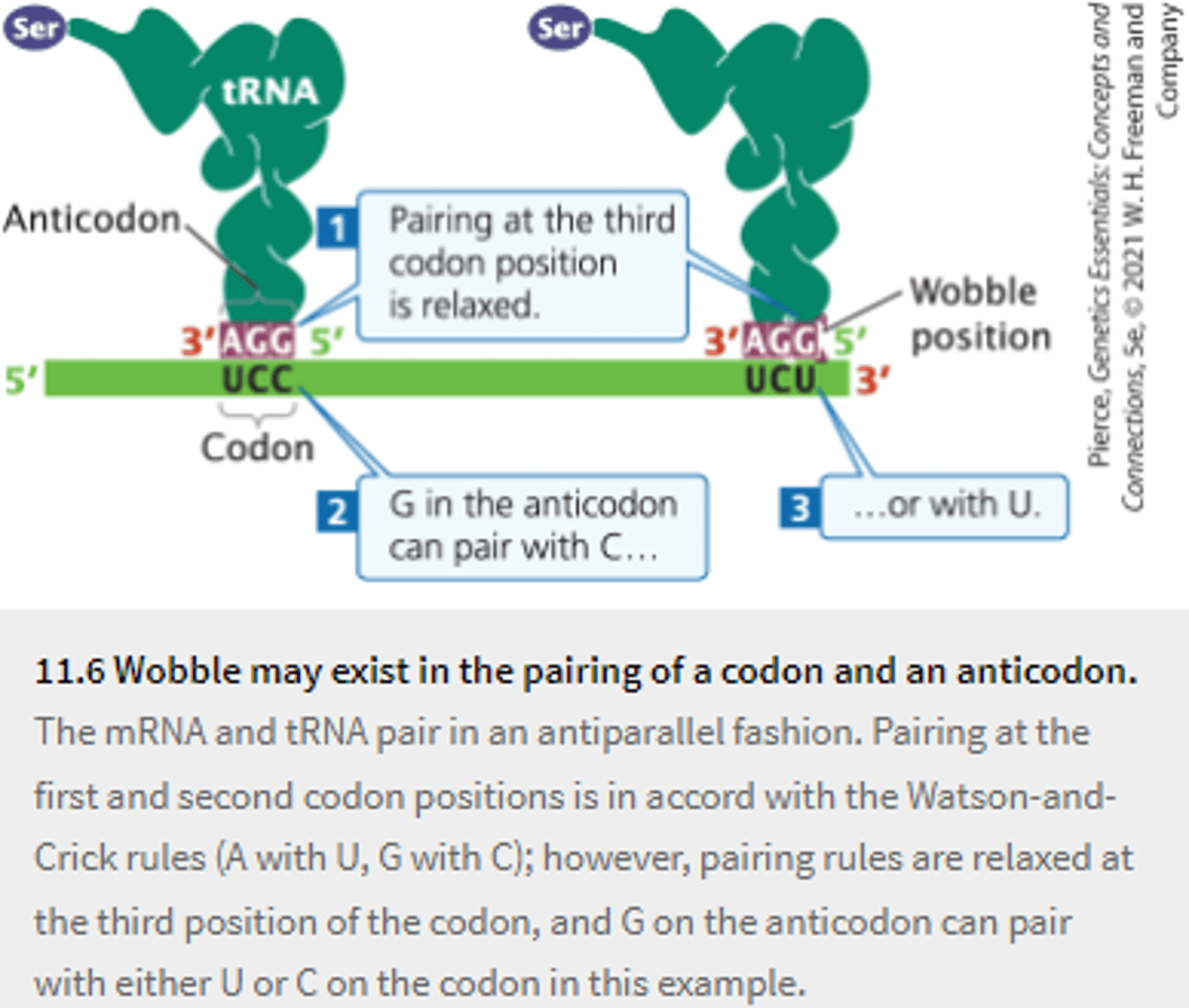
Though 61 different codons must be translated into amino acids, organisms never have 61 different tRNAs; E. coli has about 40. How can these 40 tRNAs recognize all 61 sense codons?
Wobble
Isoaccepting
Degeneracy
Redundancy
Wobble
How does the E.coli ribosome get positioned in the right place to start translation?
rRNA base-pairing with part of mRNA
Protein binding to mRNA
Binding to Kozak sequence
Assistance of poly-A tail
rRNA base- pairing with part of mRNA
Structure of RNA
polymer of nucleotides each consisting of a sugar, a phosphate group, and a nitrogenous base joined by phosphodiester bonds
-ribose sugars
-free hydroxyl (OH) group on the 2' carbon atom of the ribose sugar
-less stable than DNA
-pyrimidine uracil instead of thymine
-typically single stranded
Addition of the 5' cap
one type of modification of eukaryotic pre-mRNA
-cap is formed by addition of an extra guanine nucleotide to the 5' end of the mRNA and CH3 to the base of the guanine (a 7-methyl-guanine)
-facilitates binding of ribosome to 5' end of mRNA, increases mRNA stability, enhances RNA splicing
-cap binding proteins recognize the cap and attach to it. a ribosome binds to these proteins and moves downstream along mRNA until start codon is reached and translation begins.
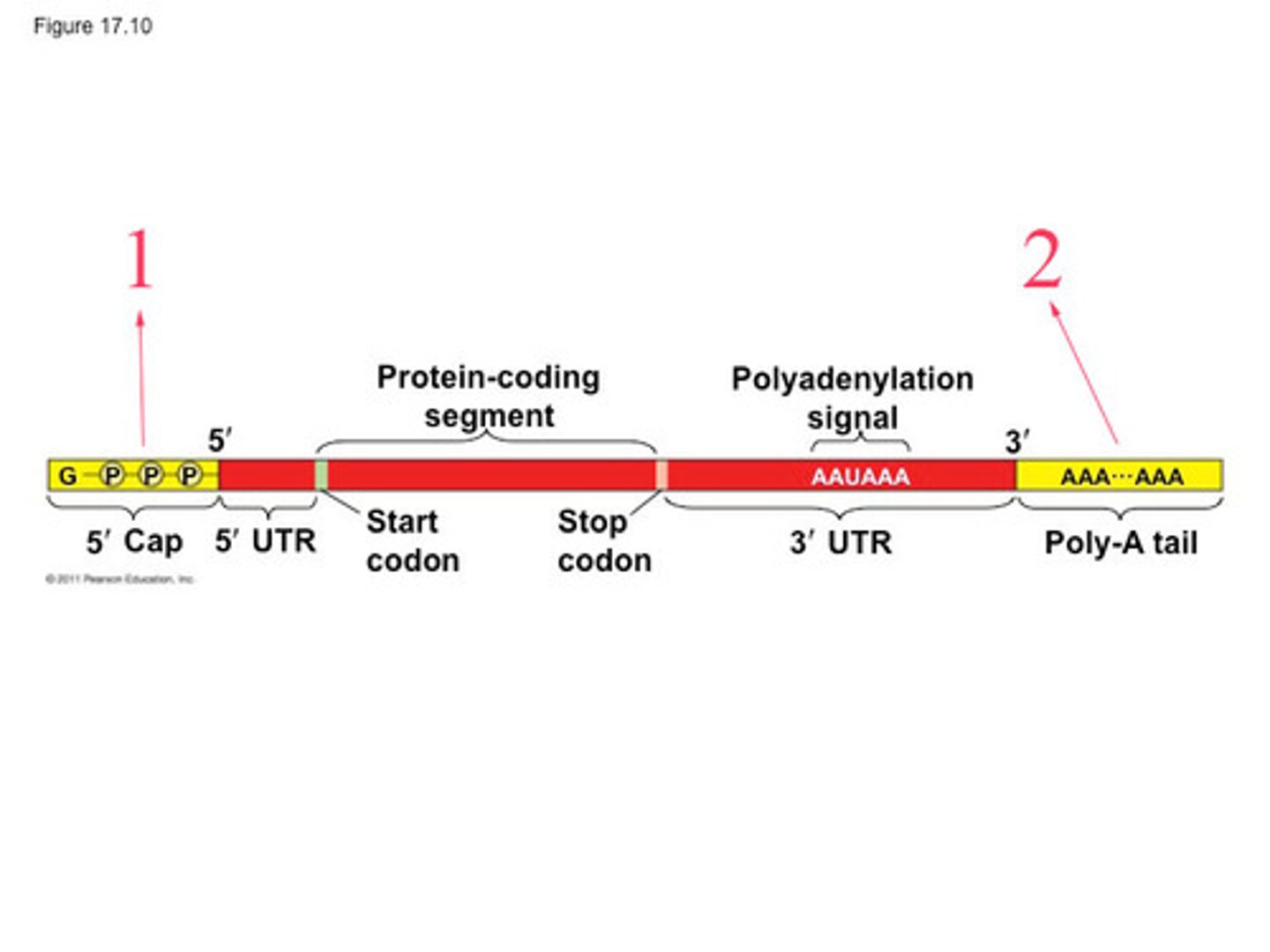
poly-A tail
Modified end of the 3' end of an mRNA molecule consisting of the addition of some 50 to 250 adenine nucleotides.
-coincident with termination of transcription
-help protect mRNA from degradation
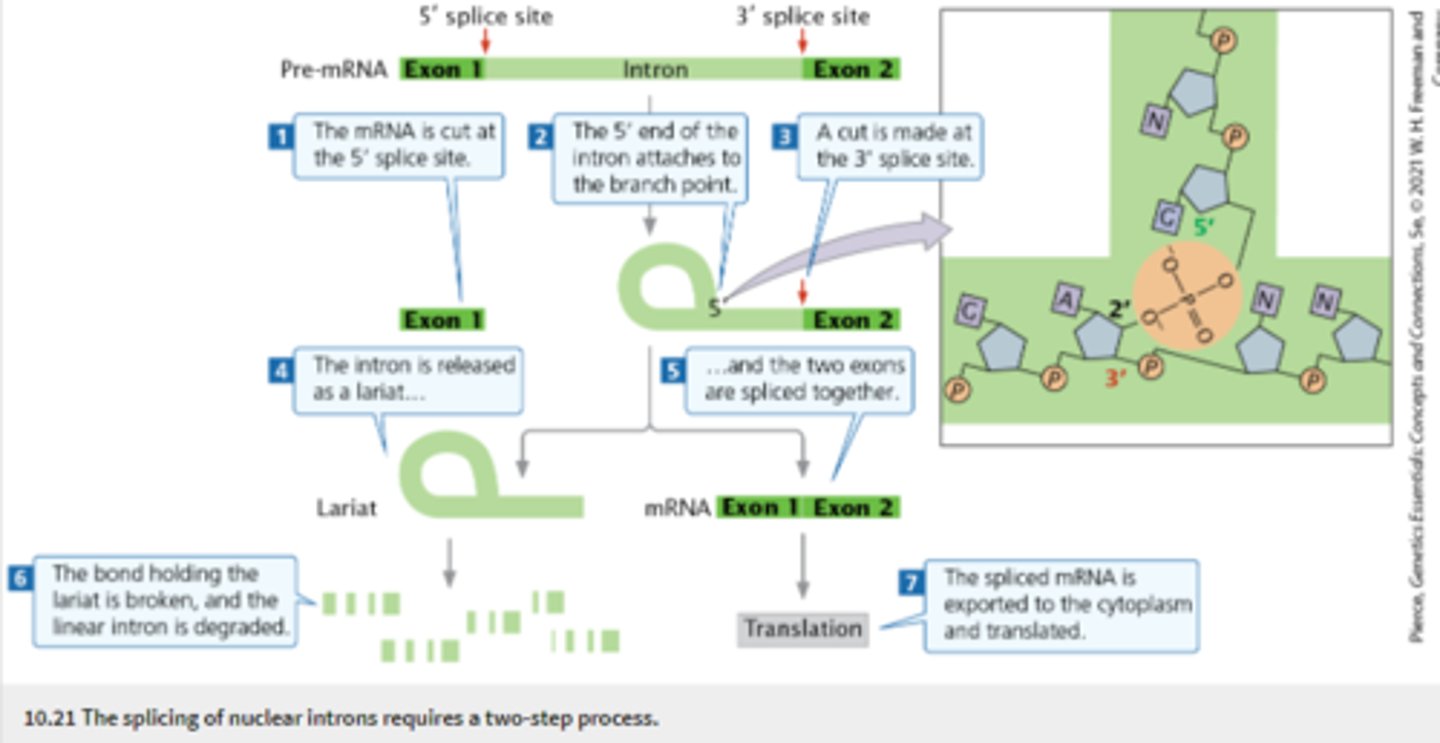
RNA Splicing
The removal of introns and joining of exons in eukaryotic RNA, forming an mRNA molecule with a continuous coding sequence
-occurs before mRNA leaves the nucleus
-takes place in spliceosome (made up of snRNPs)
requires: 5' splice site, 3' splice site, branch point (adenine nucleotide that is upstream of 3' splice site)
-introns in pre-mRNA begin with GU and end with AG
-Lariat: loop like structure created in the splicing of pre-mRNA when 5' end of an intron is attached to the branch point (hydrogen bonding)... is degraded after being released
** deletion or mutation of adenine nucleotide at branch point can prevent splicing**
3 modifications to pre-mRNA
1) 5' cap
2) poly-A tail
3) removal of introns with spliceosome
snRNPs
(small nuclear ribonucleoproteins) composed of snRNA and protein molecules, recognize the splice sites, join with additional proteins to form a spliceseome
Introns
non-coding regions
-common in euk. genes but are rare in bacterial genes.
-typically longer than exons
Exons
Coding regions
-what remains in mRNA after processing/splicing
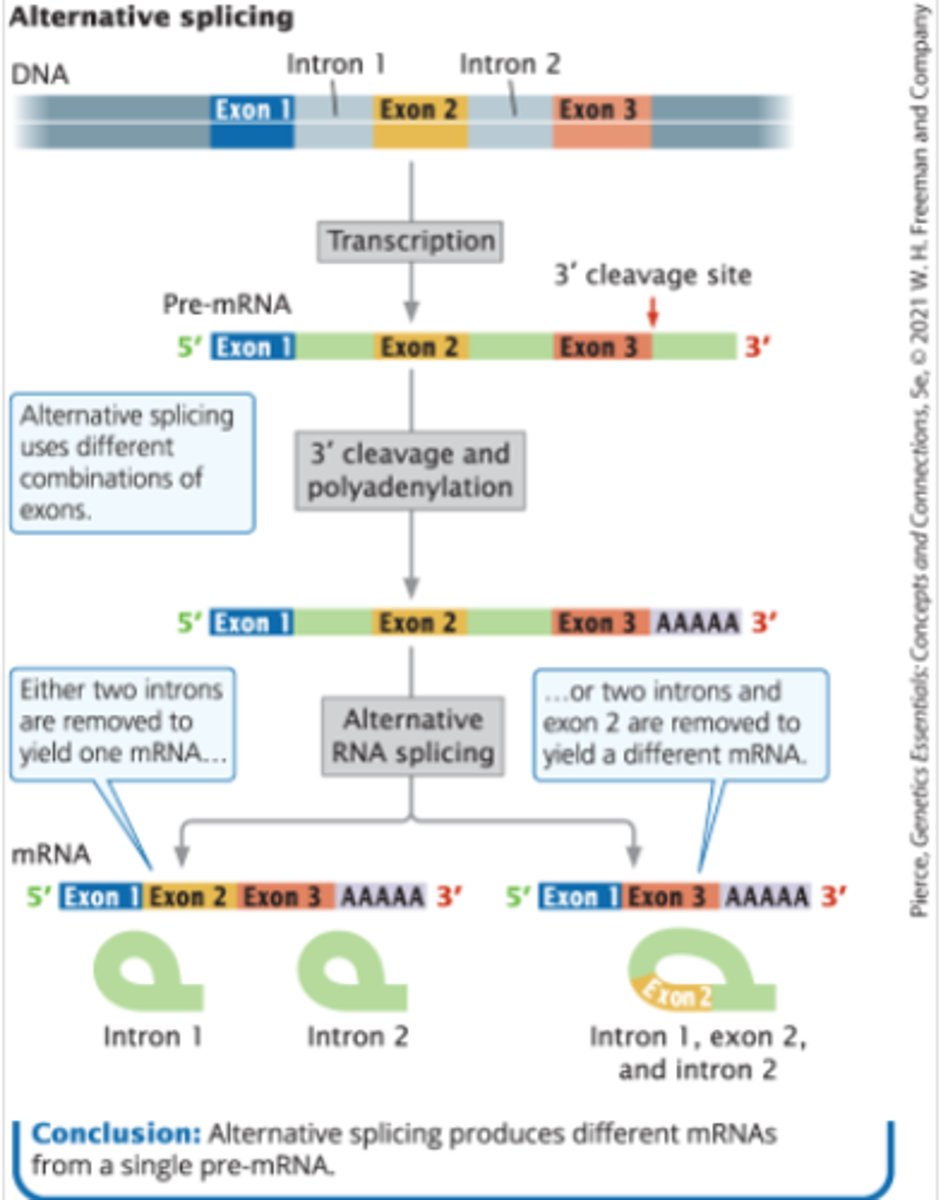
Alternative splicing
-not all exons are included in the mature mRNA
-the same pre-mRNA can be spliced in more than one way to yield different mRNAs that translate into different amino acid sequences and thus different proteins
-if there are multiple cleavage sites, the 3' exons may be excluded from the mature mRNA
ex: calcitonin pre-mRNA
tRNA processing
-several tRNAs transcribed together as one large precursor tRNA that is then cut up into pieces.
-ends are trimmed (both 5' and 3' ends)
-chemically modify bases
-addition of CCA to the 3' end by an enzyme
rRNA
ribosomal RNA; type of RNA that makes up part of the ribosome
-initially part of a long transcript, some locations are methylated, cut out, and trimmed to the right length before joining with proteins to make the ribosomes.
large ribosomal subunit and small ribosomal subunit make up ribosome
-bacteria: large 50S, small 30S.
-eukaryotes: large 60S, small 40S
mRNA
messenger RNA; type of RNA that carries instructions from DNA in the nucleus to the ribosome to code for proteins
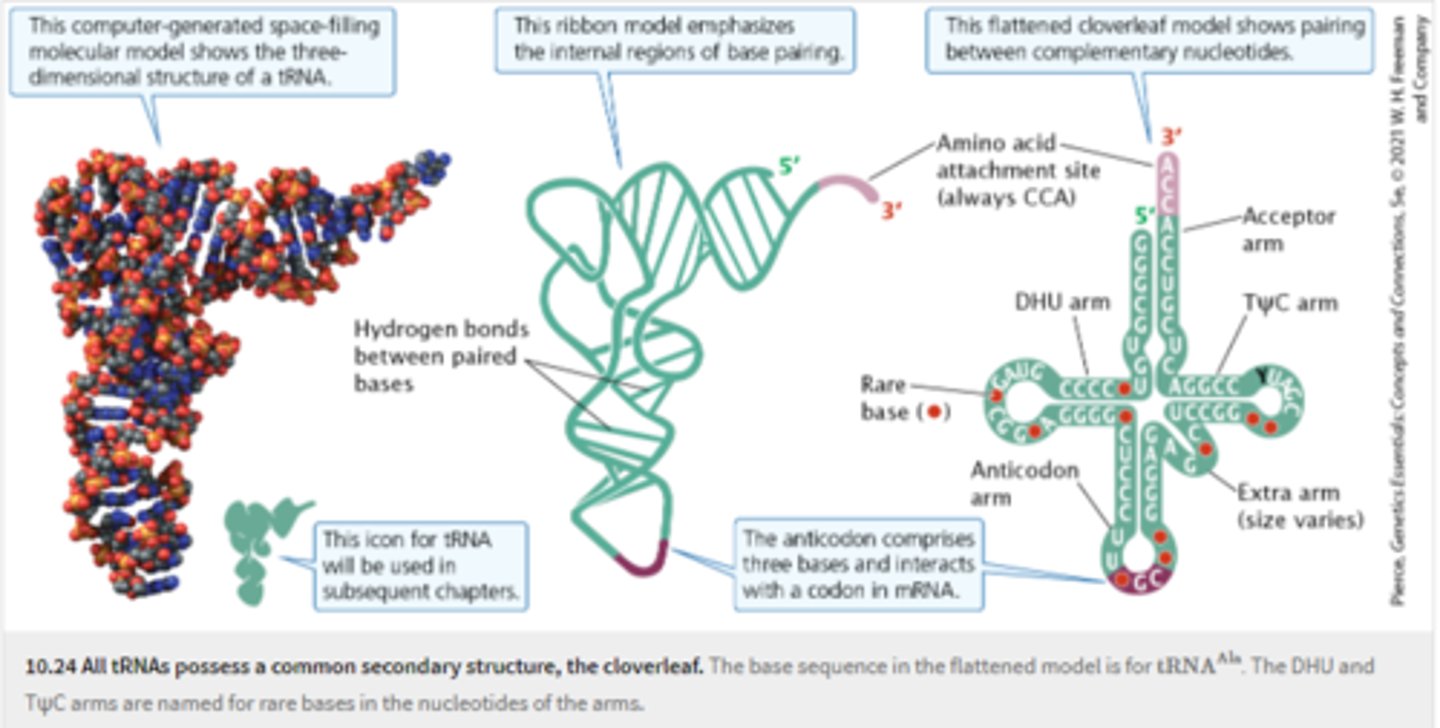
tRNA
transfer RNA; type of RNA that carries amino acids to the ribosome, incorporates amino acids into polypeptide chains
-short RNA molecules similar in secondary structure
-the same sequence, CCA, at 3' end where amino acid attaches
-3 sets of nucleotides make up anticodon
-same cloverleaf (secondary structure) folds in to L shape
link between genetic code in mRNA and the amino acids that make up a protein
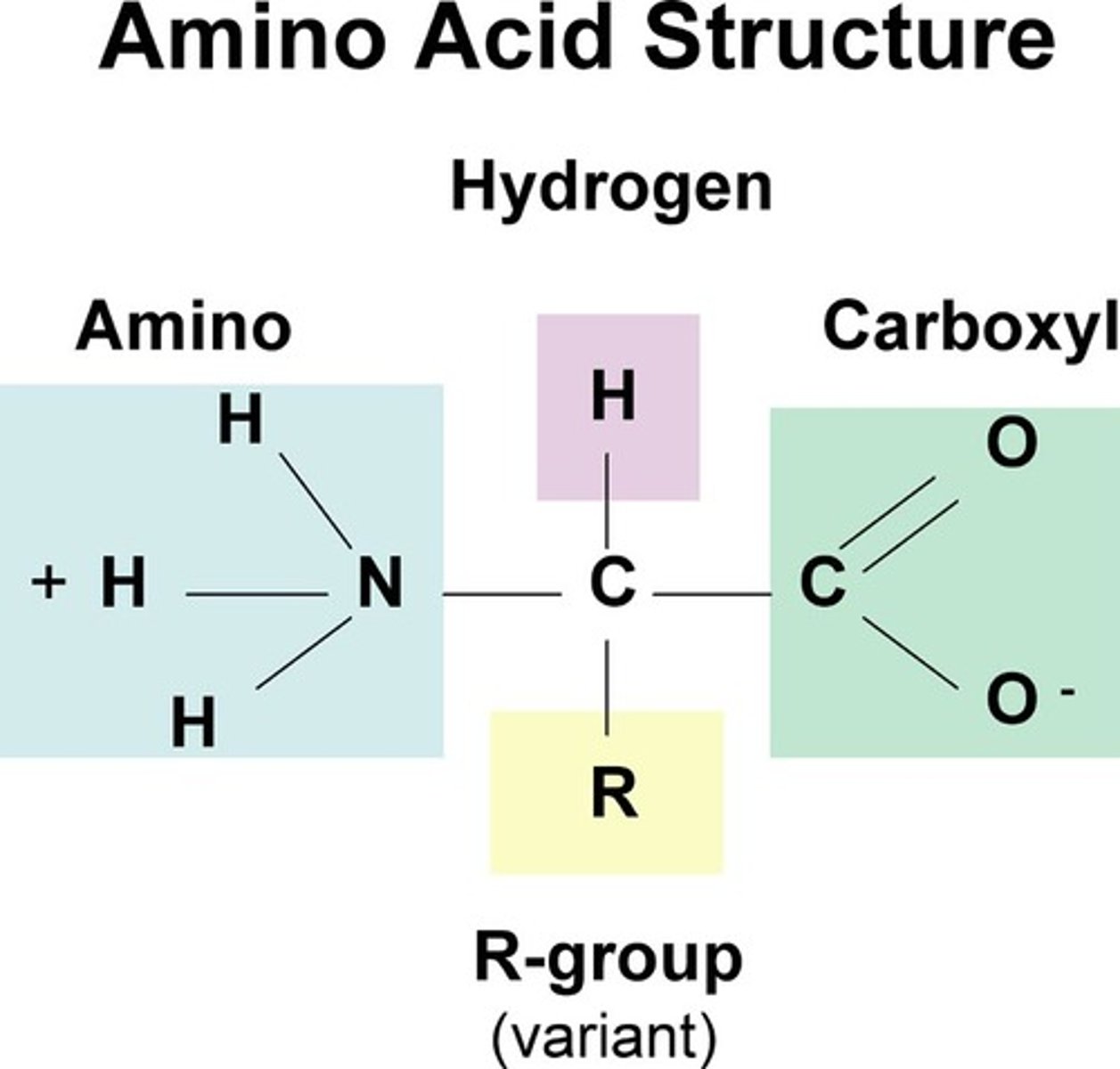
Amino Acids
central carbon atom bonded to an amino group, hydrogen atom, carboxyl group, and R group (different for each amino acid)
-monomer of proteins/ polypeptide chains
-histones are positively charged because of amino acids lysine/arginine
-Peptide bonds form between amino acids to form polypeptide chains
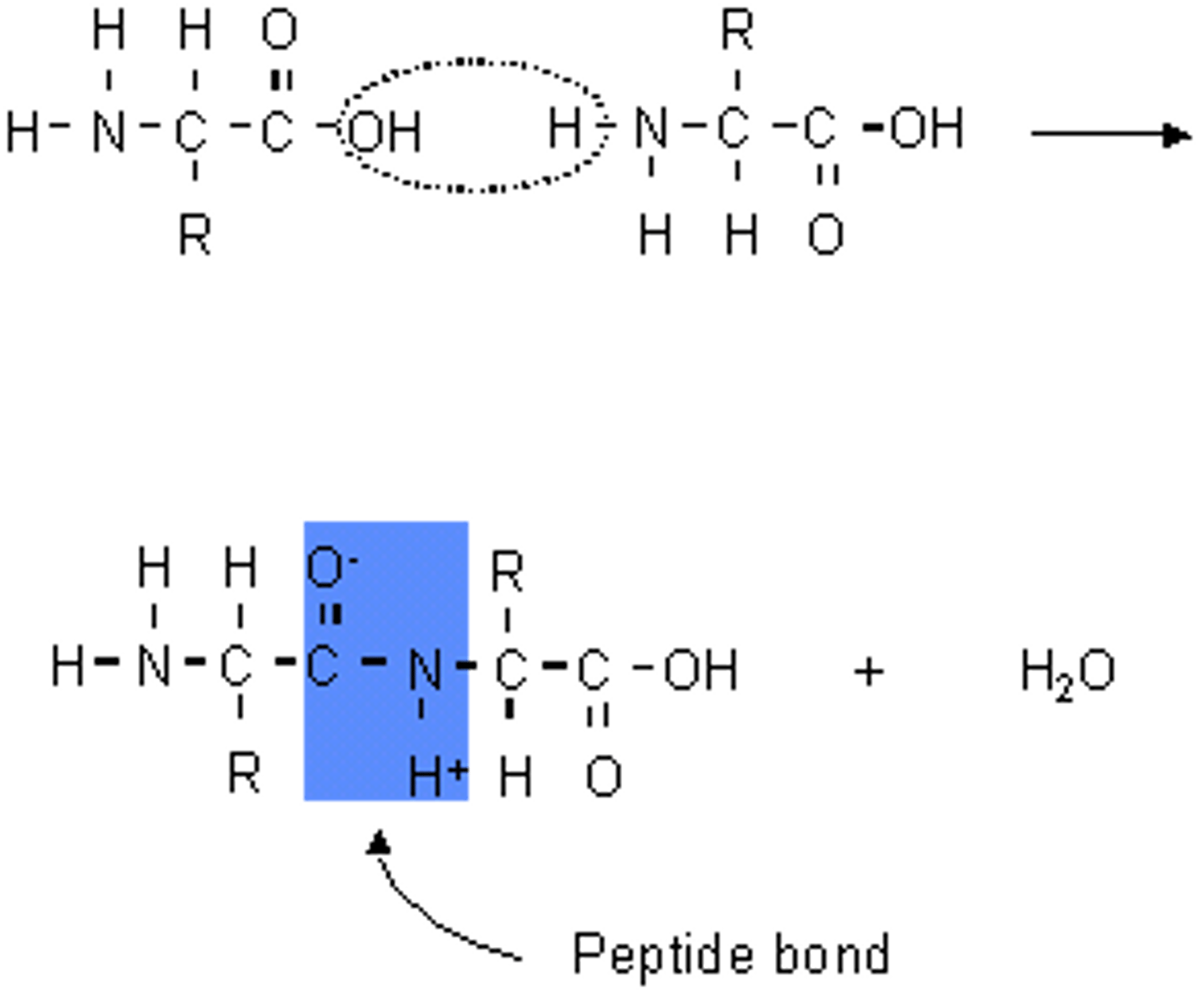
Peptide bond
The chemical bond that forms between the carboxyl group of one amino acid and the amino group of another amino acid
Transcription requires 3 major components
1. A DNA template
2. The raw materials (ribonucleotide triphosphates) needed to build a new RNA molecule
3. The transcription apparatus, consisting of the proteins necessary for catalyzing the synthesis of RNA
Codon
A specific sequence of three adjacent bases on a strand of DNA or RNA that provides genetic code information for a particular amino acid. Basic unit of genetic code
64 different codons=
3 non-sense codons code for STOP +
61 sense codons code for amino acids
Degenerate genetic code
The genetic code contains more information than is needed to specify all 20 common amino acids
-redundant, amino acids may be specified by more than one codon.
synonymous codons = codons that specify the same amino acid. (i..e UUU and UUC both specify Phe)
isoaccepting tRNAs
Different tRNAs that accept the same amino acid but have different anticodons.
-most cells have 30-50 different tRNAs, yet only 20 amino acids.
Wobble
base pairing between codon and anticodon (normally at the third, 3' position) in which there is nonstandard pairing....
allows more than one codon to pair with the same anticodon
Reading Frame
Reading mRNA nucleotides in the correct groupings.
Critically important
-genetic code is nonoverlapping, very important that code is read by the correct 3 nucleotides, set by the initiation or start codon.
Initiation codons : AUG, GUG, UUG. mark the beginning of translation and specify amino acids.
AUG is by far the most common (remember, ay u!! lets go!!
Stop codons/termination codons/nonsense codons: mark the end of translation (UAA, UAG, UGA).. these do NOT encode any amino acids.
**trinucleotide insertions (3 nucleotide insertions) do not affect the reading frame
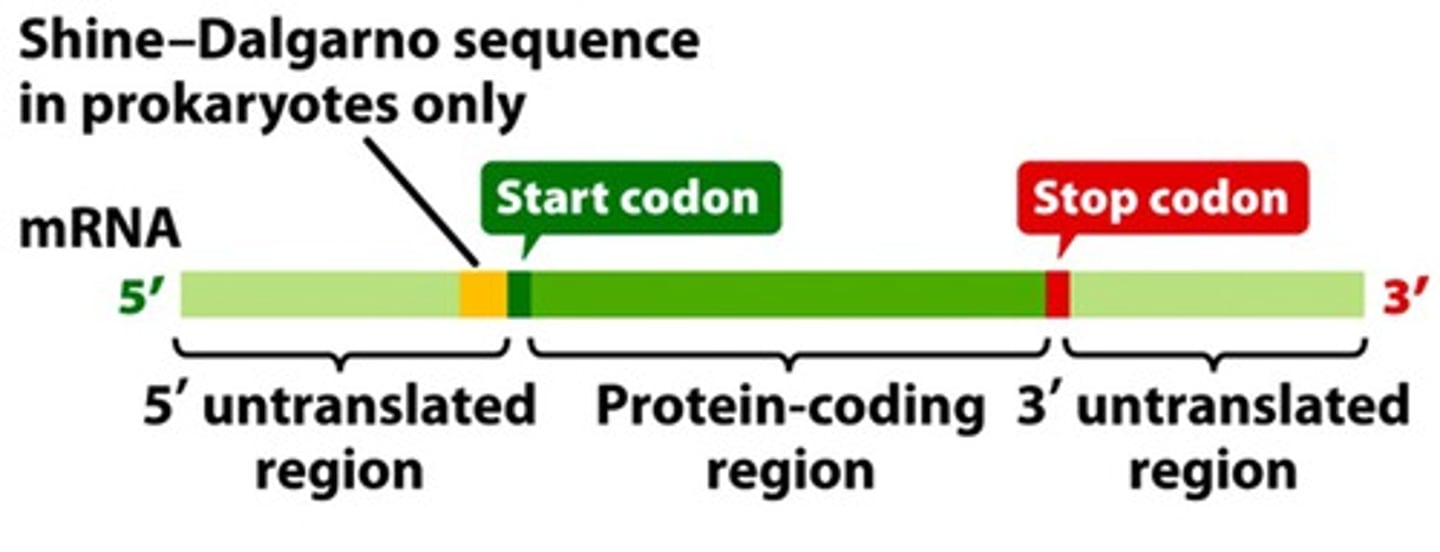
Shine-Dalgarno sequence
Serves as a binding site for ribosomes in bacterial RNA. Consensus sequence of AGGAGG.
part of 5' untranslated region
Initiation of Translation
1) mRNA binds to small subunit of ribosome
2) initiator tRNA binds to the mRNA through base pairing between initiation codon and anticodon
3) large ribosomal subunit joins the initiation complex at the P site
initiation complex moves along the mRNA until it finds the first AUG codon and begins translation
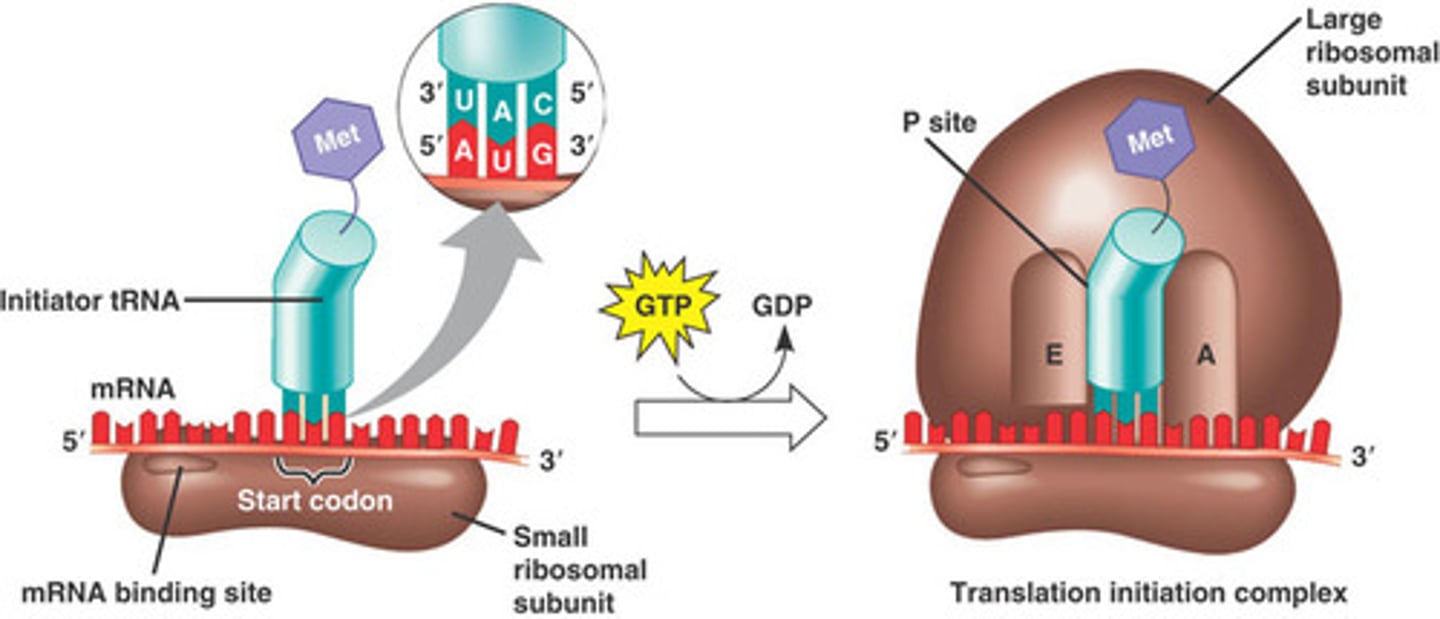
A, P, and E sites
A= aminoacyl site, think Arrival site, where new tRNAs arrive
P = peptidyl site, occupied by the initiator tRNA fMET immediately at beginning
E = exit site
Peptidyl transferase activity, a function of 23S RNA, in large ribosomal subunit, creates peptide bonds between amino acids in p and a sites.
Elongation
requires translocation of ribosome along mRNA (ribosome moves down 3 nucleotides on mRNA)
the tRNA begins in the cytoplasm -> A site -> P site -> E site -> cytoplasm.
(only exception is the initiator tRNA which begins at the P site and never occupies the A site)
-charged tRNA and its amino acid occupy the A site, a peptide bond is formed between the amino acids in the A and P sites, ad the ribosome translocates to the next codon.
Termination
translation stops when the ribosome translocates to the termination codon.
There are no tRNAs with anticodons complementary to the termination codons, so no tRNA enters the A site of the ribosome. Instead, release factors bind to the A site and promotes cleavage.
Structural genes
encode proteins that are used in metabolism or biosynthesis. typically always on
i.e. topoisomerase, enzymes of the krebs cycle
Regulatory genes
RNA or proteins interact with other DNA sequence and affect the transcription or translation of those sequences
i.e. we don't constantly need lactase or alcohol dehydrogenase
-control the expression of many of the structural genes
Operon
In bacteria - related genes are controlled together
A group of related genes transcribed from a single promotor. The basic unit of transcriptional control in bacteria.
Lac operon
Lactose - disaccharide (linkage of glucose and galactose)
Permease (LacY)- allows lactose from environment to come into cell (Y+ dominant to Y-)
Allolactose - isomer of lactose that binds to regulatory protein (produced by LacI) which makes it inactive. Because the regulatory protein can not bind to the operator, transcription of rest of lac operon occurs.
Lac I - regulatory gene with its own promotor, PI. Exerts negative control. (I+ versus I- versus Is)
β-Galactosidase (LacZ) - changes lactose to isomer allolactose and also breaks down lactose into glucose and galactose (Z+ dominant to Z-)
Transacetylase (LacA) - part of lac operon, but function unknown.
Promoter- where RNA polymerase binds to start transcription downstream (P+ dominant to P-)
Operator- includes part of the promotor, is downstream from it. Where the I protein (repressor) binds. (O+ versus Oc, where Oc means constitutive)
Operon is normally off, but can be turned on. Is inducible.
Transcription never fully stops so if a little lactose gets in to the cell through permease, it can be turned into allolactose by β-Galactosidase and then the full on transcription can begin.
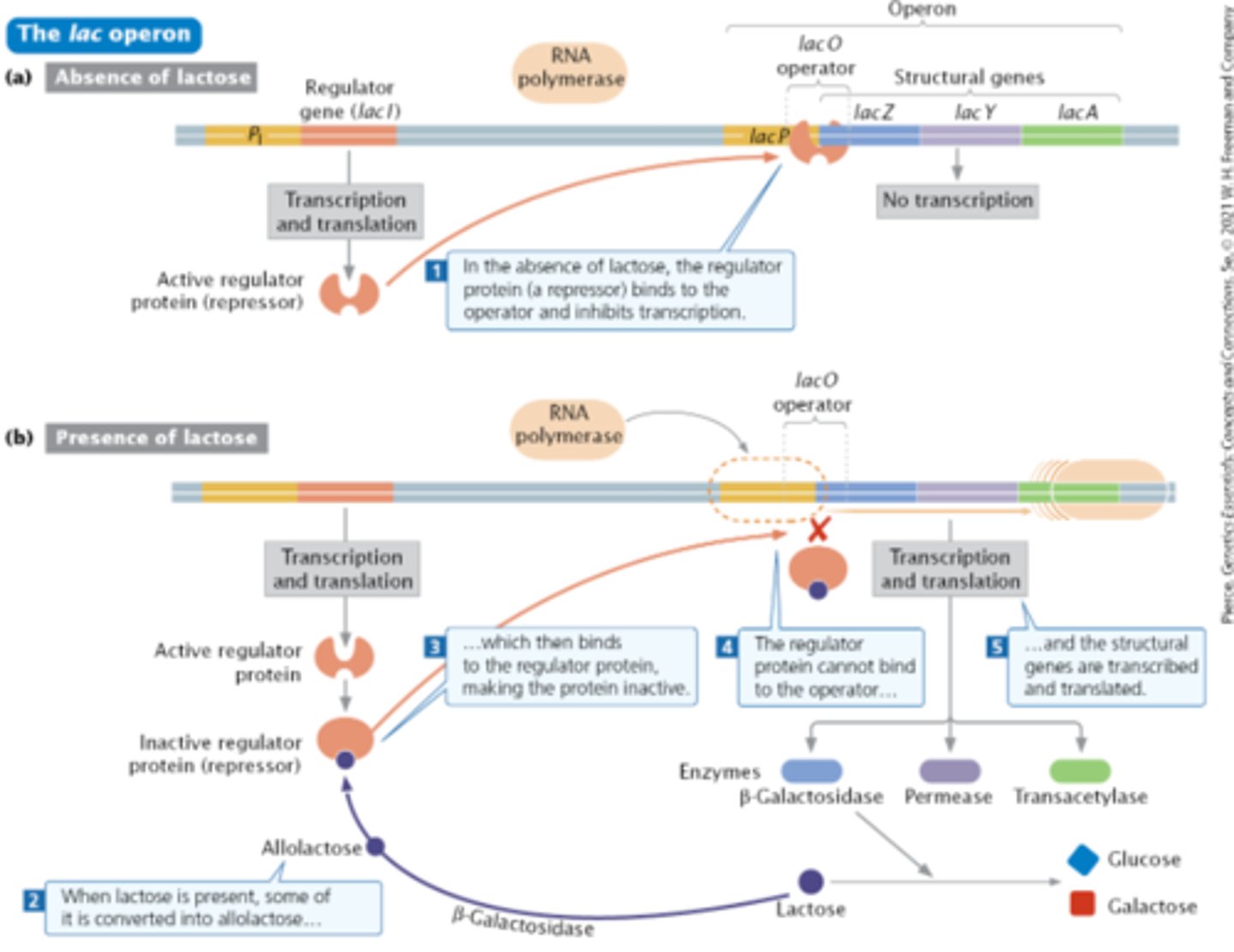
LacI
regulator gene. Produces I protein (repressor) which in the absence of lactose, binds to the operator (downstream end of Lac promotor) and inhibits transcription by blocking the binding of RNA polymerase.
Exerts negative control
I+ =dominant, normal wild type produces I protein
I- =I protein is not produced
Is =superrepressor, I protein is produced, but allolactose can not bind to it
Partial Diploids
Jacob and Monod
analyzed mutations that affected lactose metabolism.
-partial-diploid strains of E. coli include two different DNA molecules: the full bacterial chromosome and an extra piece of DNA (done through conjugation - direct transfer of genetic material between 2 cells) Created via fertility factor, F.
-F' plasmid conjugates and new partial diploid has 2 lac operons
-figured out which genes were dominant, cis acting, or trans acting
i.e. Lac- phenotypes could have genotypes LacZ-, LacY-
cis acting = able to control expression of genes only on same piece of DNA
trans acting = able to control expression of genes on other DNA (i.e. lac I protein acts on lac operon)
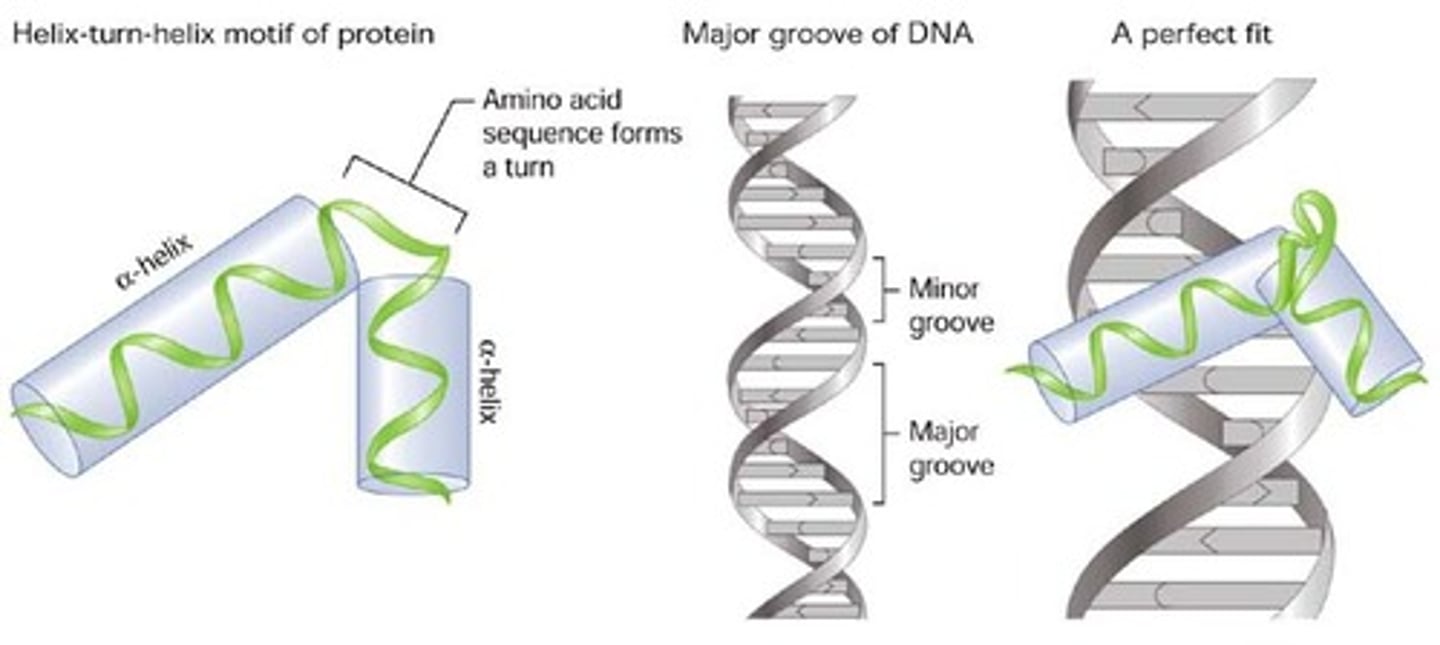
helix-turn-helix
A transcription factor DNA-binding domain in which three or four helical regions contact DNA.
β-Galactosidase
coded by the LacZ gene.
enzyme that hydrolyzes lactose to form glucose and galactose (so it can be more effectively used as an energy source)
can also isomerize lactose to allolactose
Positive Control
A regulatory protein is an activator, stimulating transcription.
-moreso eukaryotes than prokaryotes
can greatly influence the rate of transcription (i.e cAMP/CAP binding, transcription factors)
Negative Control
A regulatory protein is a repressor, binding to DNA and inhibiting transcription.
-moreso prokaryotes than eukaryotes
(ex: Lac operon, the I protein)
Constitutive Operon
-operon containing mutations and are incapable of being turned off (repressible system)
Inducible operon
Transcription is normally off, but can be turned on
(ex: Lac operon)
repressible operon
Transcription is normally on, but can be turned off.
CAP (catabolite activator protein)
RNA polymerase does not bind efficiently to many promoters unless CAP is first bound to the DNA.
cAMP/CAP make helix turn helix structure.
High glucose in cell, low cAMP. RNA polymerase does not bind well to the lac promotor.
Low glucose in cell, high cAMP. RNA polymerase binds really well to the lac promotor. Causes maximum rate of transcription of lac operon.
trp Operon
tryptophan binds to the repressor protein and enables it to repress gene transcription. (aporepresser becomes activated by corepressor)
tryptophan = essential amino acid.
If cell is present in an environment with a lot of trp, it will just import it instead of making it.
Example of a repressible operon (i.e. it is always on, but can be turned off).
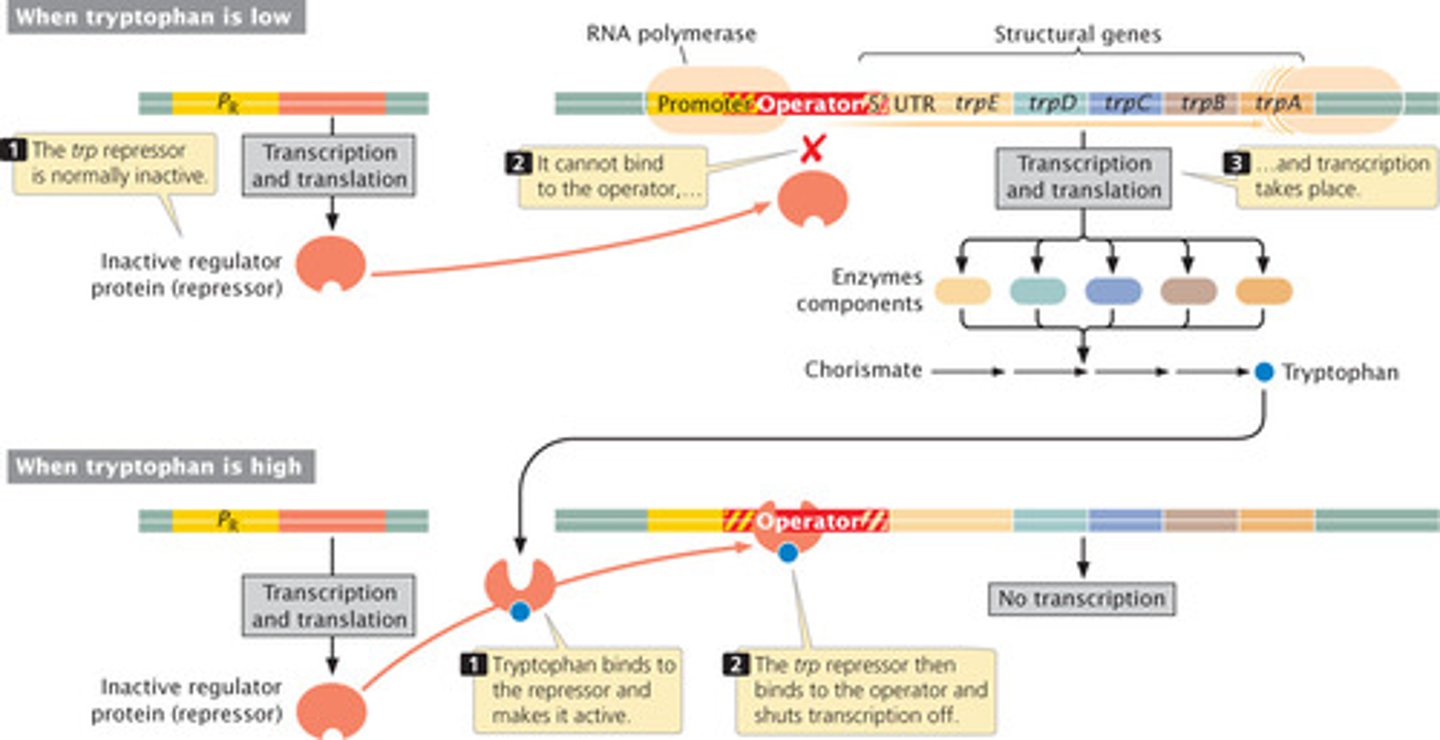
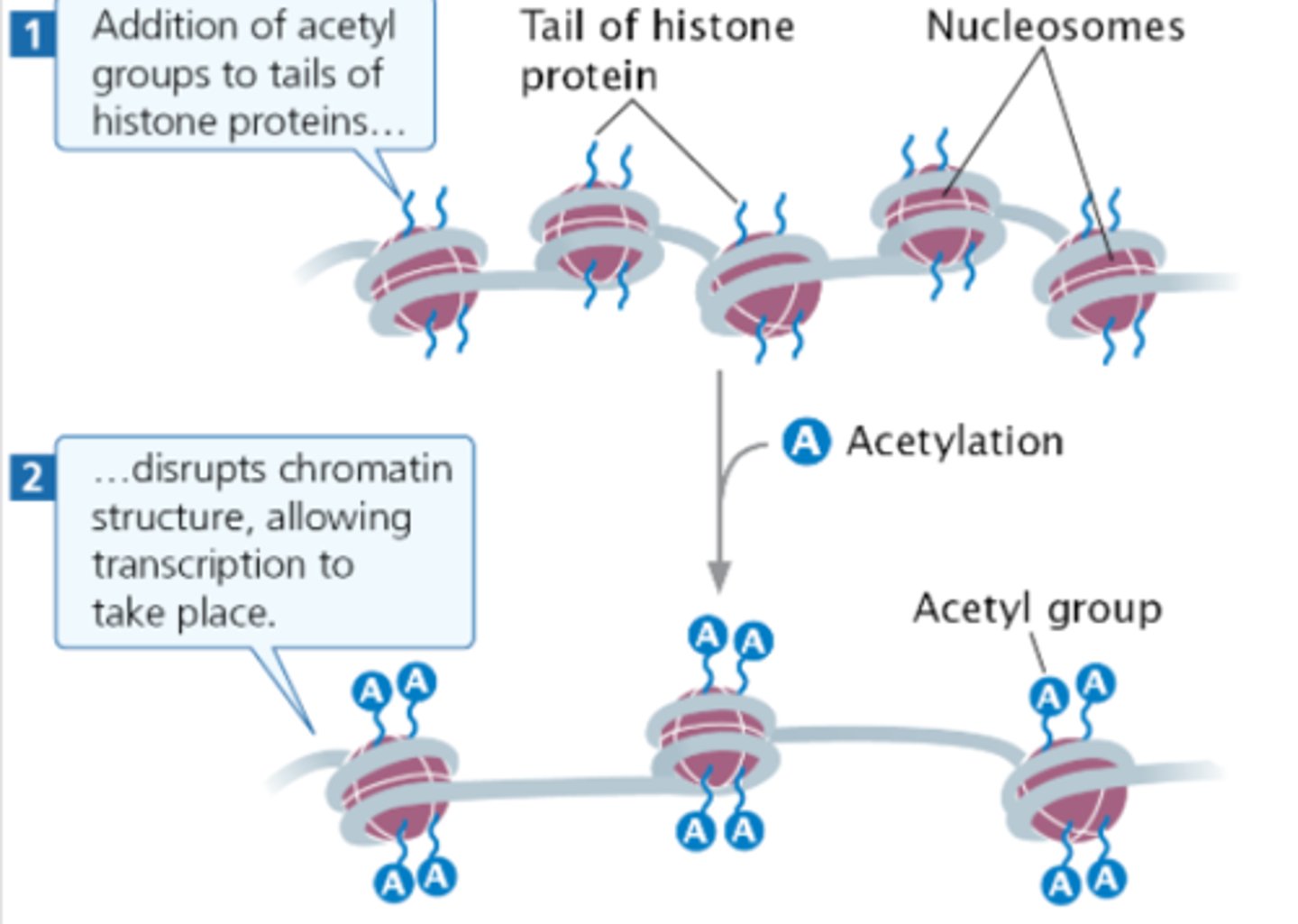
Histone acetylation
DNA is normally wrapped around histones. These DNA fragments are unable to be translated and transcribed.
Adding an acetyl group causes the DNA to bind less tightly so it can be translated and then transcribed.
Transcription factors
Collection of proteins that mediate the binding of RNA polymerase and the initiation of transcription.
By itself, the basal transcription apparatus performs transcription very slowly.
With transcription factors (transcriptional activating proteins) binding to enhancer sequences, the rate of transcription in eukaryotes can be greatly increased.
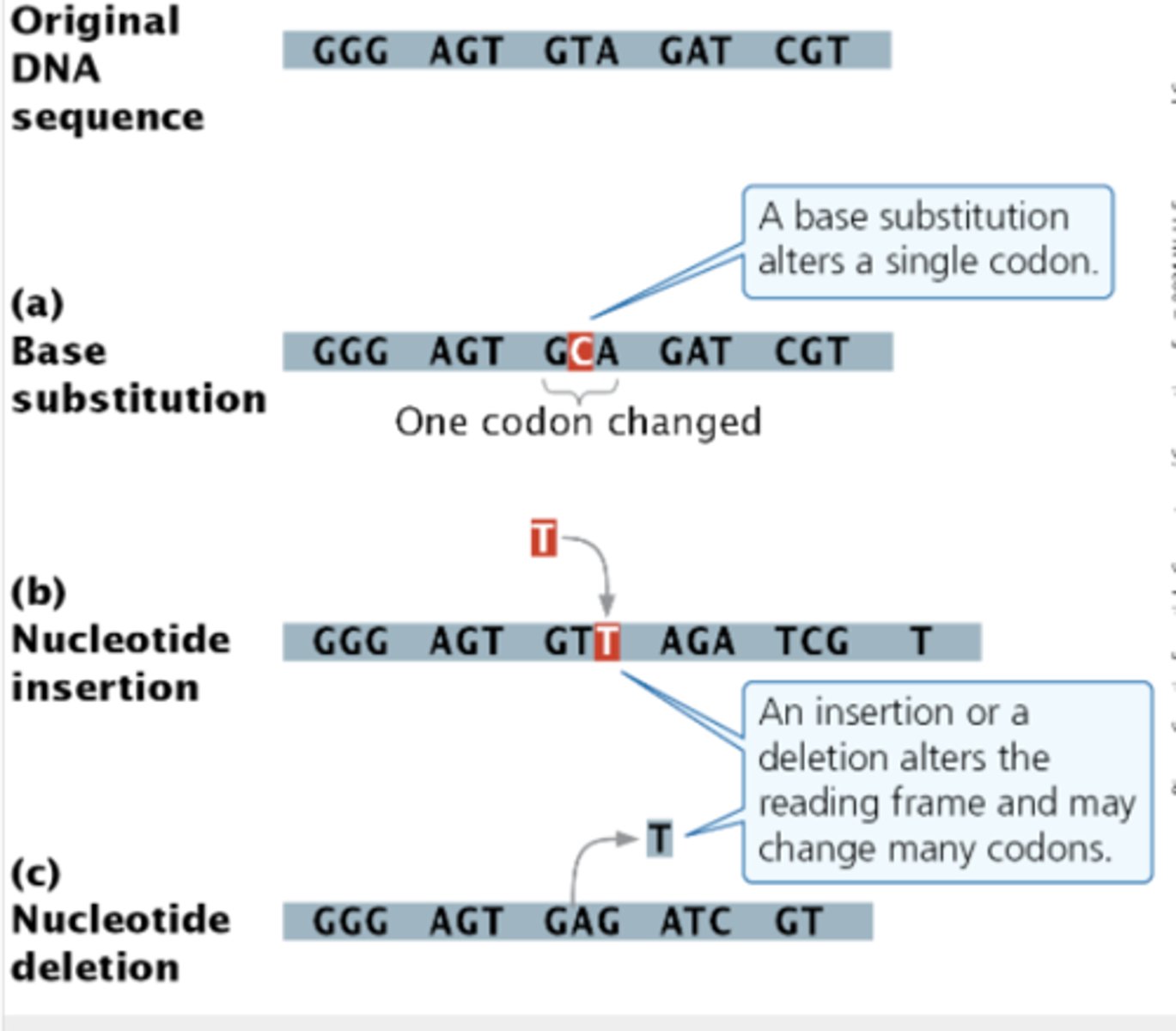
Mutations
Inherited change in the DNA sequence of genetic information.
Somatic mutations - arise in somatic tissues, divides my mitosis, mutation is passed on to clone daughter cells, many have no phenotypic effect overall
Germ-line mutations- arise in cells that produce gametes, passed on to offspring and has a much more pronounced effect
Substitution
Deletion
Insertion
Inversion (very rare at the level of gene)
Change in promotor sequence of an operon would affect
how tightly RNA polymerase binds to the sigma factor, would affect transcription
5' untranslated region
sequence of nucleotides at the 5' end of mRNA; does not encode the amino acids of a protein
3' untranslated region
sequence of nucleotides at the 3' end of mRNA; does not encode the amino acids of a protein but affects both the stability of the mRNA and its translation
Substitution
1) change amino acid (diff codon), nonsynonymous
2) does not change amino acid, synonymous
3) creates a stop codon, non-sense change
Insertions/Deletions
change in reading frame, very serious effect
Phenotype consequences to mutations
-none
-loss of function (mendel's wrinkled seeds)
-gain of function (constitutive expression of FGFR3, leads to achondroplasia (dwarfism))
Huntington's disease
Genetic disorder that causes progressive deterioration of brain cells.
trinucleotide repeats CAGCAGCAGCAG. # of repeats increase over time. reading frame is stable, but high frequency of same amino acid can cause misfold.
We all have this gene, but < 37 repeats = normal,
> 37 repeats = neurodegenerative disease
Gain of function, dominant, mis-folding protein
Frequency of mutation
Normally, very rare but depends on measure.
Ex: Achondroplasia is 4 in 100,000 births, very rare because rate of substitution per base pair is very rare.
BUT
lactose intolerance, is much more common because Lac- could mean I-, LacZ-, LacY-, etc. more opportunities for one mutation to make the gene inoperable.
Human parents normally give their offspring around 50 new mutations (not normally seen because coding genome is only 1-2% of the 6.5 billion bp)
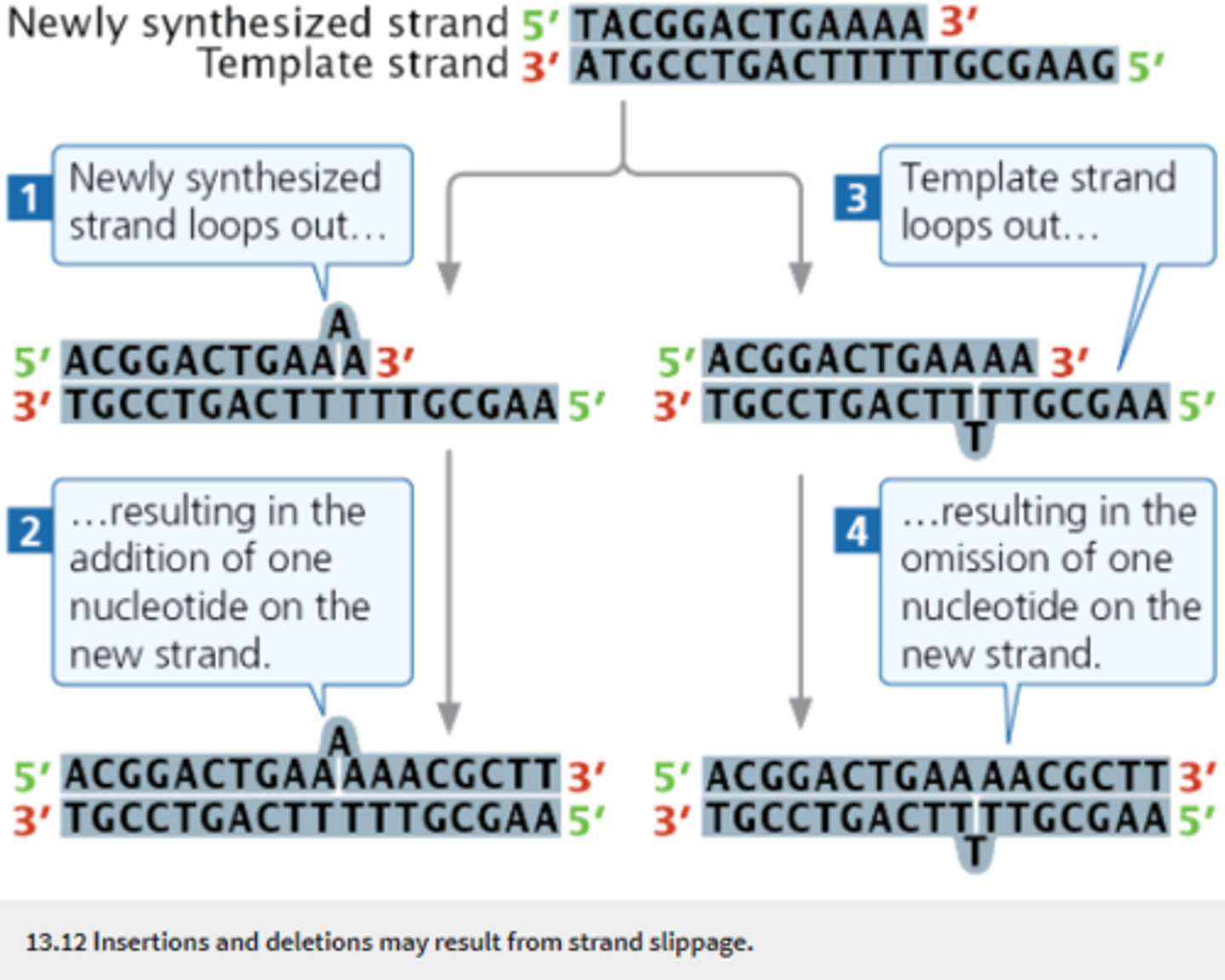
Strand-slippage
Slipping of the template and newly synthesized strands in replication in which one of the strands loops out from the other and nucleotides are inserted or deleted on the newly synthesized strand.
Loss-of-function mutation
causes the complete or partial absence of normal protein function (i.e. mendel's wrinkly seeds)