Genetics I: Nucleic Acids and Chromatin
1/29
Earn XP
Description and Tags
» Explain the structure and function of DNA as the genetic material. » Describe the different types of genetic information contained within DNA. » Illustrate how DNA is packaged into nucleosomes, chromatin, and chromosomes. » Define and discuss chromatin remodeling and epigenetic regulation, and explain their impact on gene expression
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
describe the flow of genetic information
DNA → RNA → Protein
DNA is replicated, then transcribed to RNA, then translated into a protein
just read this quickly

describe the structure of DNA
buidling blocks are nucleotides
base pairing rules: A ←> T (2 H bonds), C-G (3 hydrogen bonds)
Double-stranded DNA are antiparallel:
one strand: 5’ → 3’
another strand: 3’ → 5’
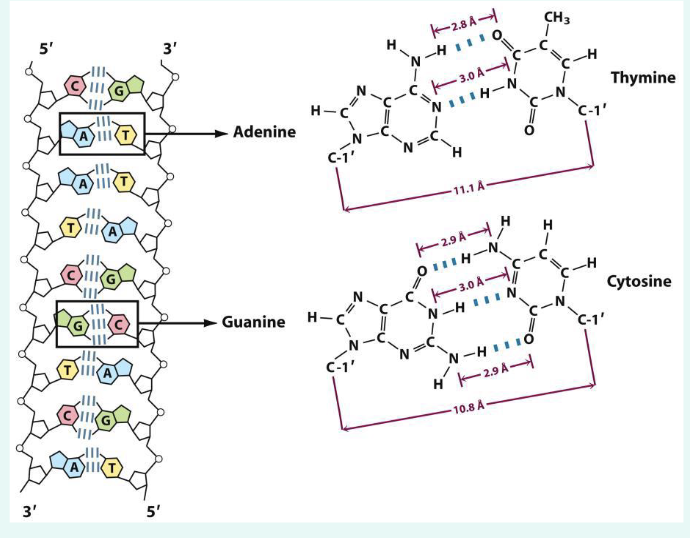
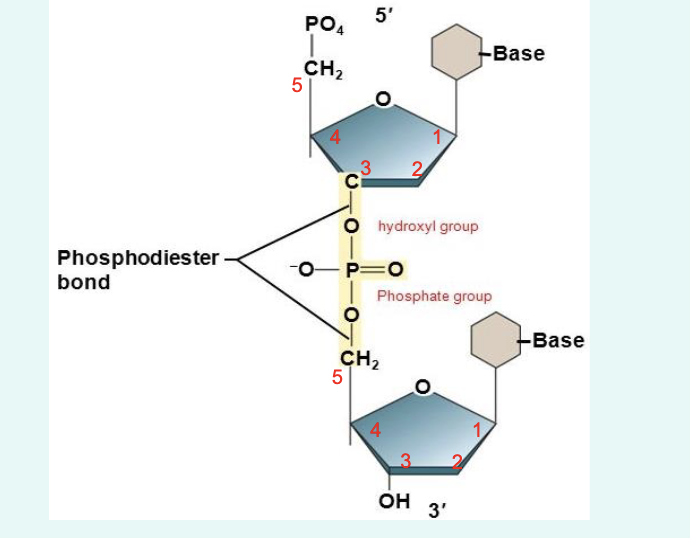
what are phosphodiester bonds
» Chemical bonds that link two nucleotides to form the
backbone of a DNA strand
» Hydroxyl group (–OH) on 3’ carbon of one sugar
joins phosphate group (–PO4) on the 5’ carbon of the
next nucleotide to form a phosphodiester bond
» 5’ – phosphate group
» 3’ – hydroxyl group of a pentos
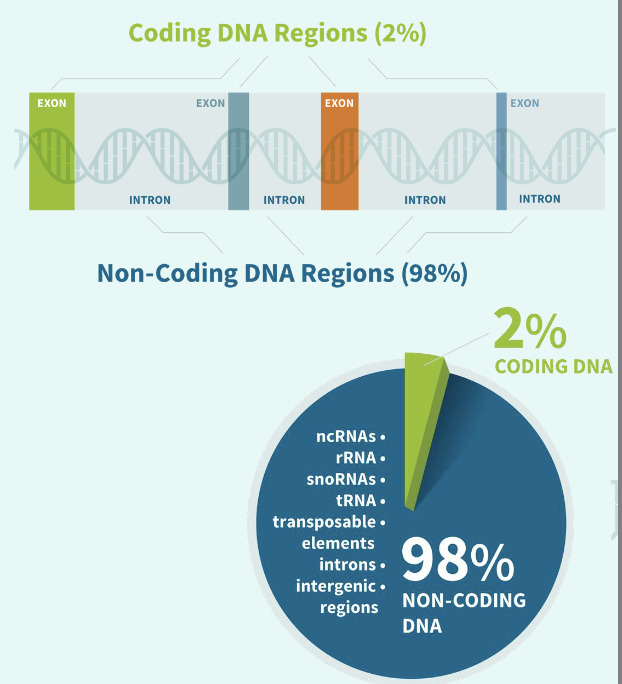
how many base pairs in human genome?
around 3.2 billion - human genome project
how many chromosomes
23 pairs of chromosomes - 2 pairs → 46 in total
how many base pairs of DNA in one cell?
6 billion base pairs
how many base pairs are used for encoding protein?
only 2% - protein coding genes
98% are “junk” - are you wasting energy to produce DNA that are mostly useless?
cells contain the same genetic material (DNA) true or false?
true
so smth else makes them develop into diff cell types…
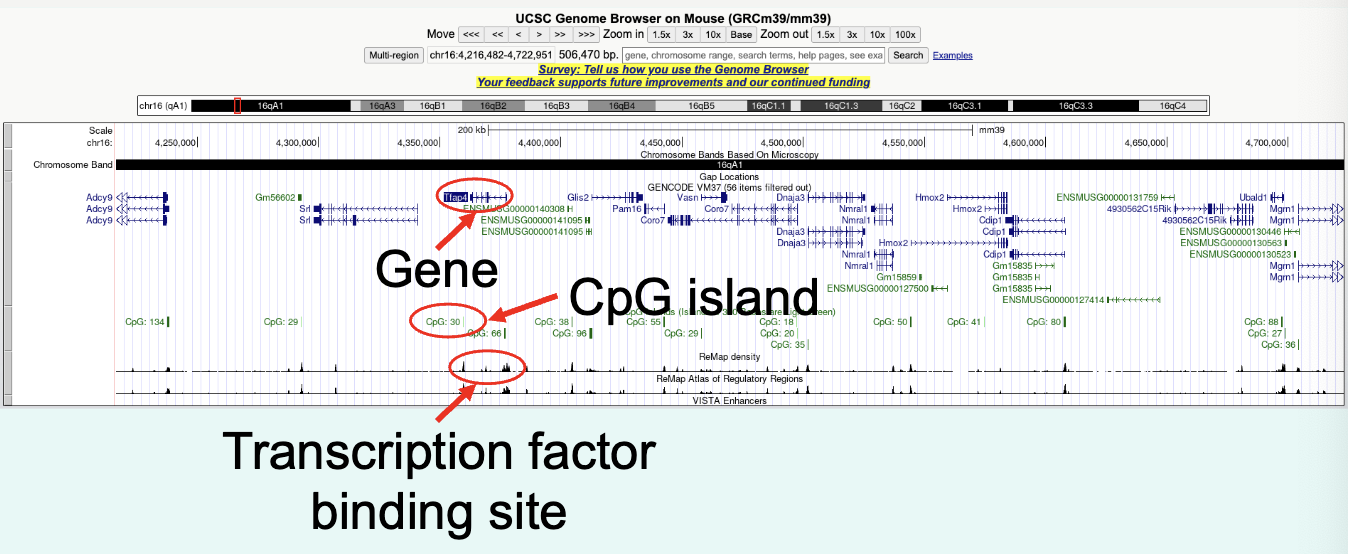
what info is encoded within DNA?
gene, CpG island, transcription factor binding site
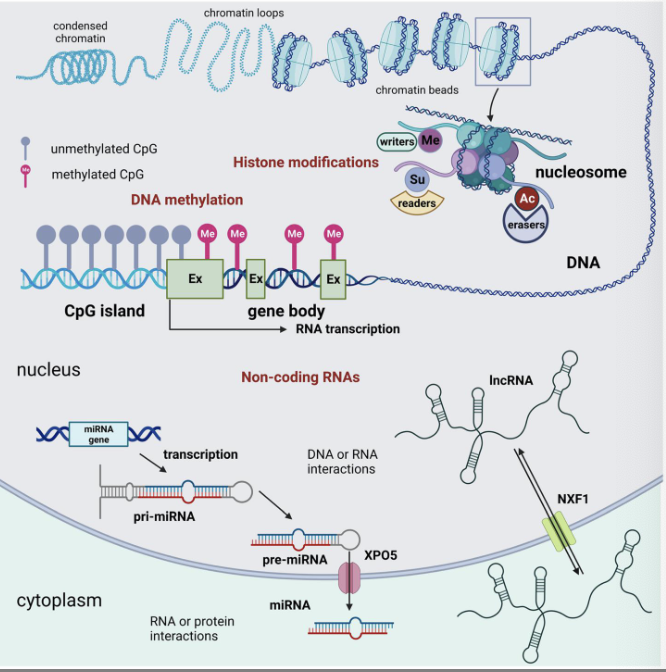
“junk” DNA are actually useful - so what do they
» ”Junk” DNA has regulatory roles
» CpG island: DNA methylation
» Methylated CpG: less accessible
» Unmethylated CpG: more open → transcription
machinery can bind → genes are more active!
» Histone modifications: open/closed chromatin
» Non-coding RNAs: stability/activity of mRNA
» All affect DNA accessibility/stability
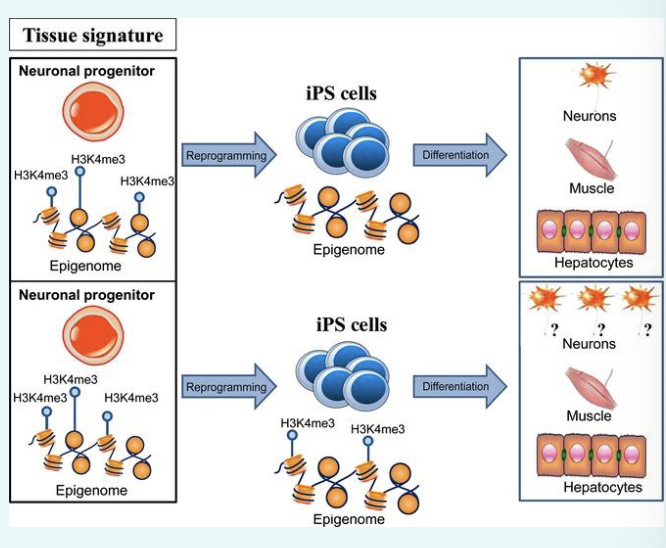
what is epigenetic memory?
how cells “remember” their identity
epigenetic modifications allow individual cells to maintain a specific gene expression pattern
such modifications can be copied during DNA replication
what are chromosomes?
» Densely packed structure of DNA and associated proteins
→ usually appeared during cell division
» DNA wrapped around histone → nucleosomes → chromatin
(interphase; non-dividing stage) → chromosome (condensed
dividing stage)
» Sister chromatids: identical copies (chromatids) formed during
DNA replication, joined by centromere
» Gene: Unit of genetics – a length of DNA that contains the coding
sequence for RNA/protein.
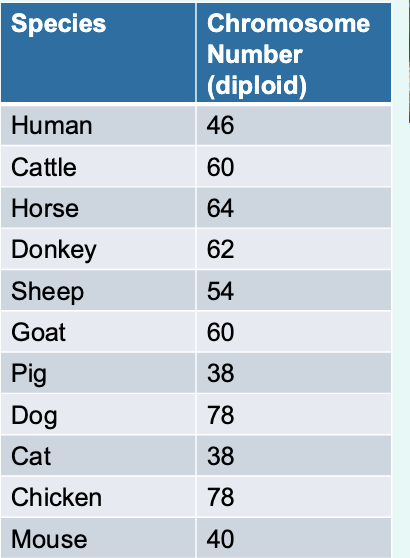
» The number of chromosomes varies depending on the species
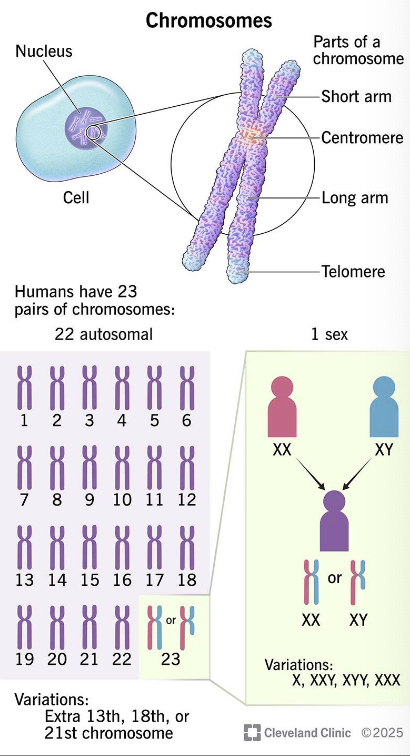
look at the chromosome number (diploid) for species
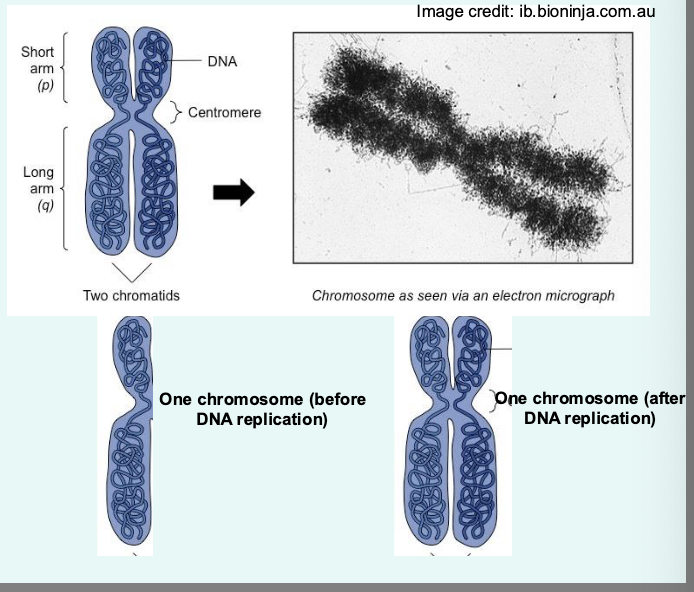
describe the architecture of a chromosome
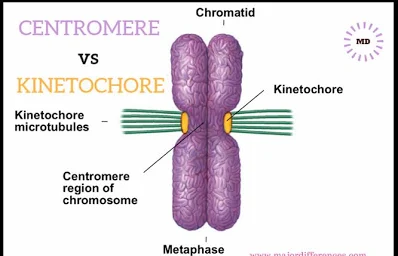
centromere: links a pair of sister chromatids together during cell division → kinetochore attach to this structure for chromosome segregation
composed of short (p) and long (q) arm: divided by constriction point called centromere
useful when mapping the location of genes/mutations within the chromosome
telomere: ends of the chromosome: repetitive DNA sequences: shorten during aging
Hayflick limit: the number of times a normal somatic cell can divide before cellular sensescence (stop dividing)
critically short telomere trigers DNA damage responses and cellular senscence → cell death
what is a somatic cell?
any biological cell forming the body of a multicellular organism that is not a reproductive cell (gamete)
what is kinetochore?
(multi-protein complex composed of mainly microtubules)
summarise DNA vs Chromosome
DNA
» Molecule that stores genetic information
» Made of nucleotides (A, T, C, G).
» Exists as a double helix.
» Serves as a template for making proteins and
provides instructions for cell function
Chromosome
» A highly organised, compact structure of DNA +
histone proteins.
» Packages long DNA molecules to fit inside the
nucleus
» Becomes highly condensed and visible during
cell division
» Contains many coding genes, regulatory
regions, and other non-coding sequences.
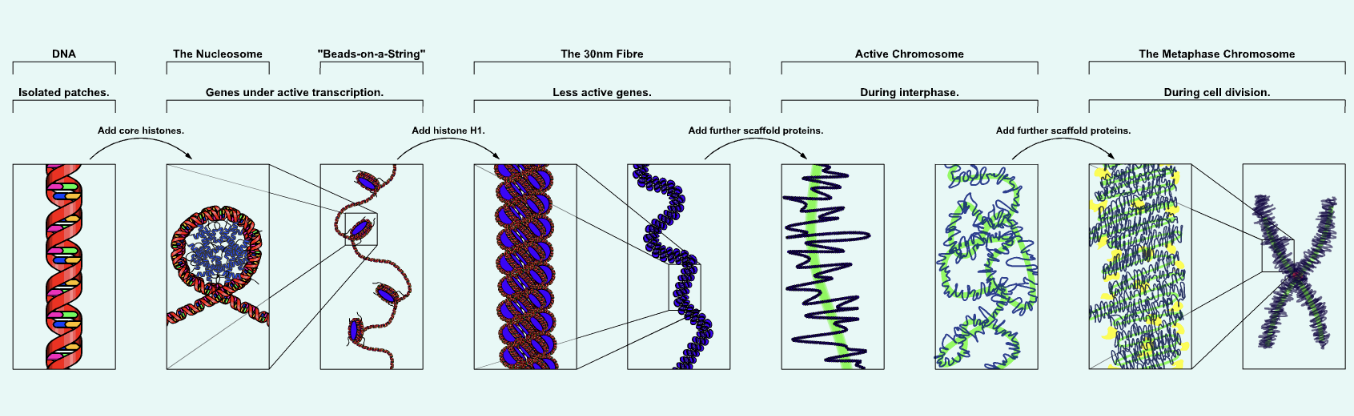
what is chromatin?
a complex of DNA + proteins packages genetic material inside the nucleus of eukaryotic cells
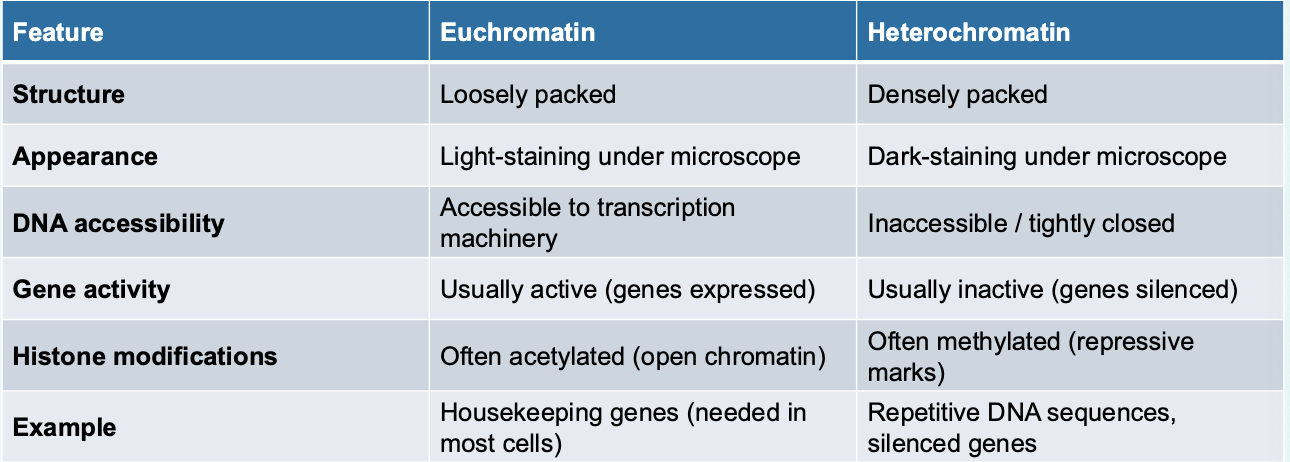
Chromatin exists in two forms: euchromatin → less condensed, can be transcribed, and heterochromatin → highly condensed; typically not transcribed
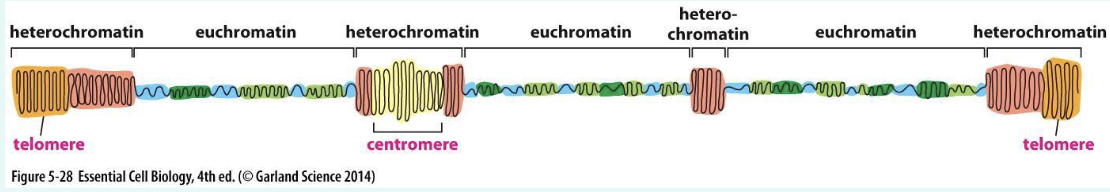
compare heterochromatin vs euchromatin
remember • Euchromatin = “loose beads on a string” → transcription factors can bind.
• Heterochromatin = “tight knot of beads” → transcription blocked
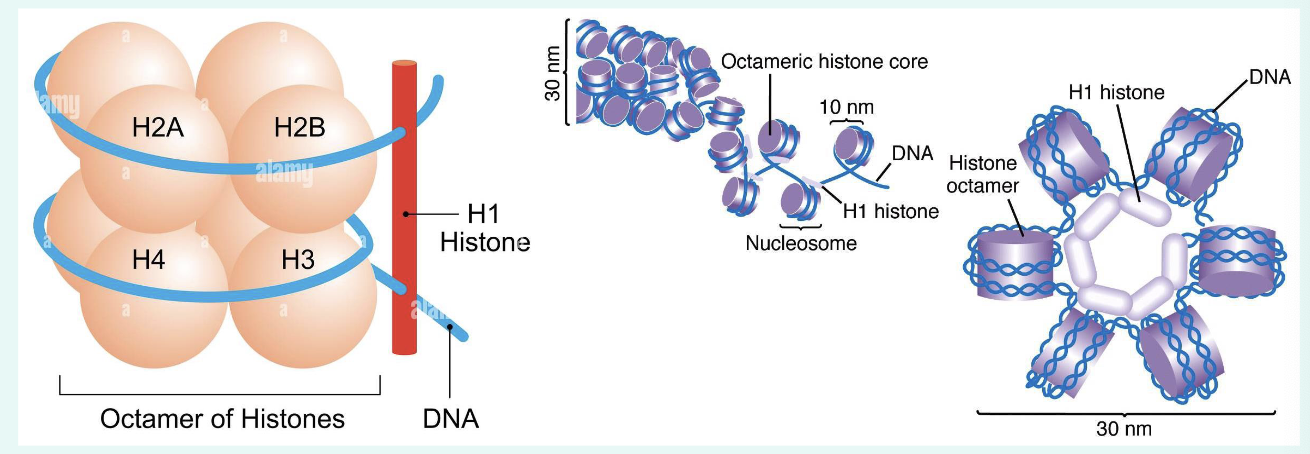
what is histone?
basic proteins (positive charged) that DNA (negatively charged) wraps around to form nucleosomes
core histone: H2A, H2B, H3 and H4 to form histone octamer
H2A-H2B Dimer + H3-H4 dimer
Histone H1: linker to compact nucleosomes
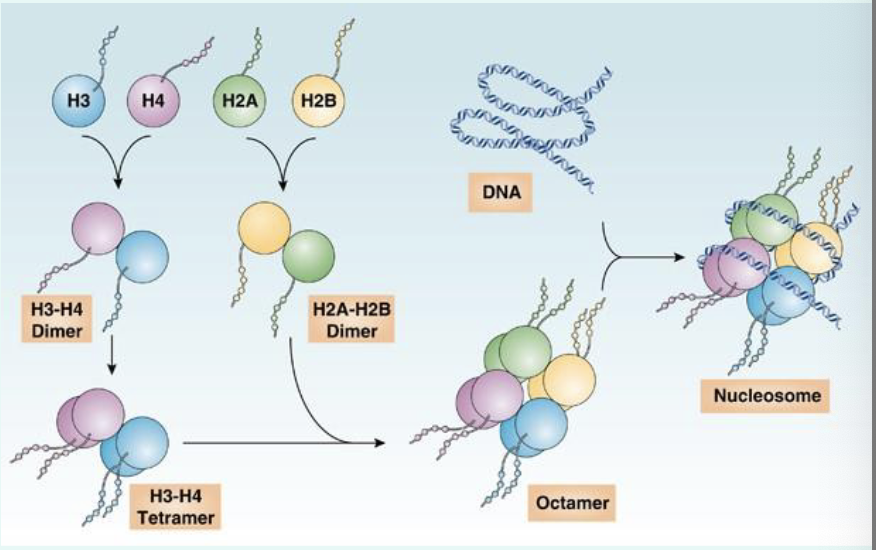
what is a nucleosome?
DNA wrapped around histone octamer; each unit of tiny “beads-on-a-string” with a diameter of approximately 11nm
less than two turns of DNA (147bp) wrapped around histone octamer
the fundamental subunit of chromatin
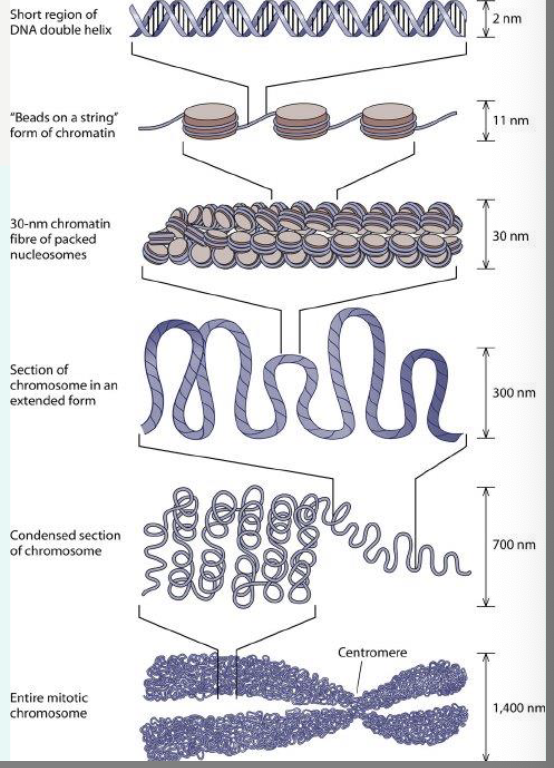
what is the chromatosome?
nucleosomes are then folded around each other to form a fibre
a chromatosome is basically histone h1 + nucleosome
chromatin fibres are then folded into loops
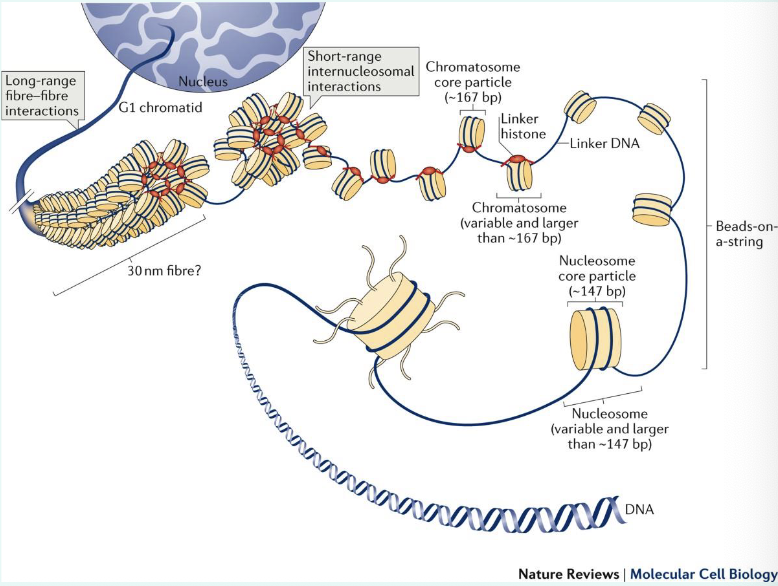
summarise DNA packaging
you’re gonna a need a video on this
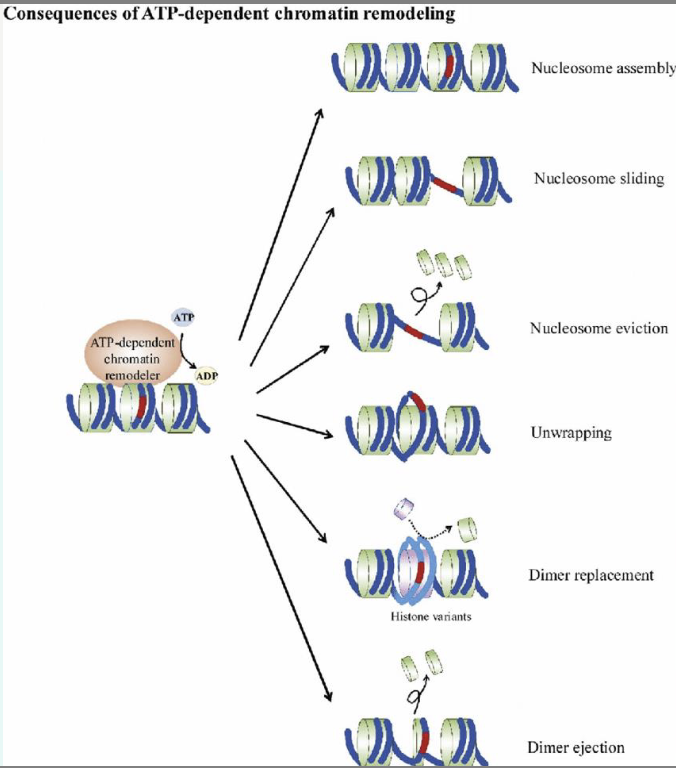
what is chromatin remodelling?
for context….
chromatin are structurally dynamics:
→ compact DNA within the nucleus (euchromatin vs heterochromatin)
→ control how and when specific genetic information is used
the actual definition…
it is the dynamic modification to control DNA accessibility for transcription, replication and repair
Why is it important to be able to control how DNA is used?
different cells in an animal have different functions - e.g. a neuron needs to make neurotransmitters, a β-cell in the pancreas needs to produce insulin
an animal has different needs throughout its life - juvanile animals need to grow
animals need to adapt to different environments
• An animal adapted to a poor diet becomes more efficient at storing energy when food is abundant.
• Animals in cold environments increase metabolism.
• Tanning – more melanin production by melanocyte
what is chromatin remodelling?
ATP-dependent chromatin remodelling complexes - move, eject or restructure the nucleosome
covalent histone modifications by specific enzymes
• Histone acetyltransferases (HATs),
deacetylases (HDAC), methyltransferases
(HMT), and kinases
• Exposes promotor sequences → transcription
machinery can be easily accessed.
• Increases the frequency of transcription
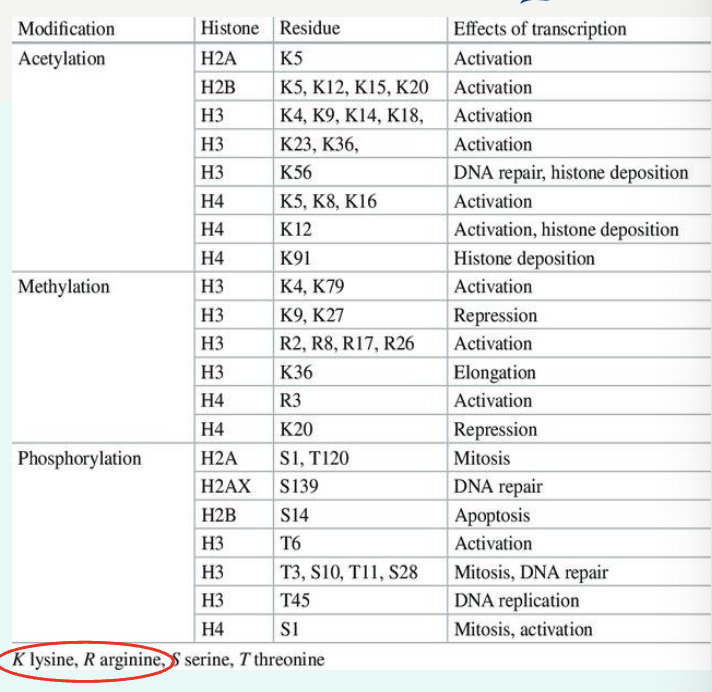
what is histone modification?
histone “tails” can be chemically modified (acetylation, methylation, phosphorylation)
→ affect gene expression
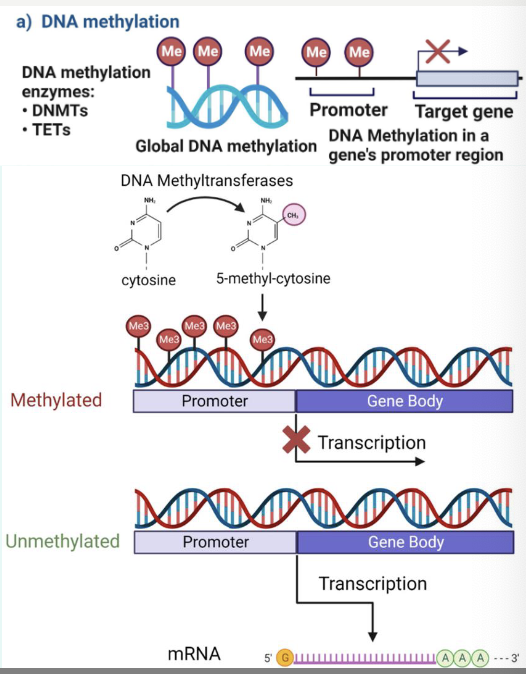
what is DNA methylation?
» Occurs in CpG sites (not necessarily CpG island)
→ add a methyl group (–CH3) to cytosine based in DNA
» CpG sites: where C is followed by G in 5’-3’ direction
» CpG island: regions of DNA rich in C–G sequences
→ Often near gene promoters
» Enzymes involved:
DNA methyltransferases (DNMTs) → add –CH3 group
TET (Ten-eleven translocation enzymes) → remove it!
» Resulted in gene silencing (or gene activation when
removed)
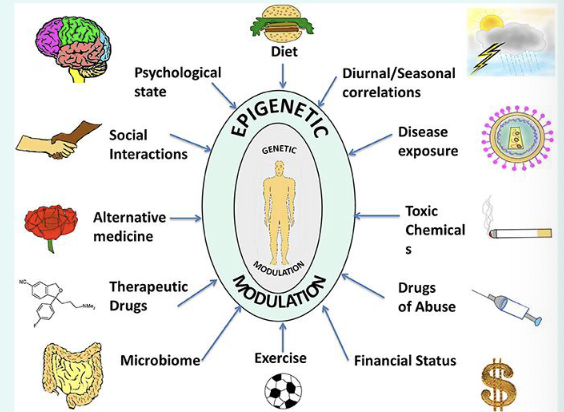
what are epigenetic effects?
» Influence the phenotypic changes by changing gene
activity without altering the genotype (DNA sequences).
» Major mechanisms:
• DNA methylation.
• Histone methylation/acetylation/phosphorylation.
• Chromatin remodelling.
» Gene: ON OFF.
» Inherited during cell division (epigenetic memory)
» Inherited to offspring if happened in germ cells:
Inherited changes without changing genetic information