BIS104 MT2 Practice Questions
1/27
Earn XP
Description and Tags
IM GOING TO $^&%$# LOSE IT
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
28 Terms
You isolated a gene that encodes the MITO120 protein. Based on the bioinformatic analysis, you predict that MITO120 will localize to the mitochondrial matrix. You then confirm the localization of MITO120 in the mitochondria using fluorescence microscopy. Where would you expect to find the fusion protein if you fuse the MITO120 protein to the C-terminus of GFP?
a. None of the above
b. Mitochondria
c. Cytoplasm
d. Nucleus
c. cytoplasm
You have isolated a gene that encodes the UNC22 protein. Based on microscopy analysis, you determine that UNC22 is a cytoplasmic protein. If you add the PKKKKRRKV peptide sequence to the middle of UNC22, where do you expect to see the fusion protein within the cell?
a. Golgi
b. ER
c. Nucleus
d. Cytoplasm
c. nucleus
In your research project, you have identified the UNC100 protein that contains the PKKKRRRKP sequence. You create a fusion of UNC100 with GFP and observe it using fluorescence microscopy. The results indicate that UNC100-GFP localizes to the nucleus. Next, you mutate two of the lysine residues to alanine (PKAARRRKP), and this fusion of the mutant UNC100 protein with GFP abolishes nuclear localization.
What would you conclude from this experiment?
a. The PKKKRRRKP sequence is neither necessary nor sufficient to drive UNC100 into the nucleus.
b. The PKKKRRRKP sequence is not necessary to drive UNC100 into the nucleus.
c. The PKKKRRRKP sequence is necessary to drive UNC100 into the nucleus.
d. The PKKKRRRKP sequence is sufficient to drive UNC100 into the nucleus.
c. The PKKKRRRKP sequence is necessary to drive UNC100 into the nucleus.
You isolated a gene that encodes MITO119 protein. From the bioinformatic analysis, you predict that MITO119 should localize to the mitochondria. Then, you confirm the localization of MITO119 to the mitochondria using fluorescence microscopy. If you fuse the mitochondrial matrix targeting sequence of MITO119 to the N-terminus of GFP, where would you expect to find GFP?
a. Nucleus
b. Cytoplasm
c. ER
d. Mitochondria
d. mitochondria
You have isolated a gene that encodes the UNC22 protein. From the microscopy analysis, you determine that UNC22 is a cytoplasmic protein. If you add the PKKKKRRKV peptide sequence to the N-terminus of UNC22, where would you expect to see the fusion protein in the cell?
a. Golgi
b. ER
c. Nucleus
d. Cytoplasm
c. nucleus
You isolated a gene that encodes the MITO19 protein. Based on the bioinformatic analysis, you predict that MITO19 should localize to the mitochondria. You confirm the localization of MITO19 to the mitochondria using fluorescence microscopy. If you insert the mitochondrial matrix targeting sequence of MITO19 into the middle of GFP, where would you expect to find GFP?
a. Mitochondria
b. Nucleus
c. Cytoplasm
d. ER
c. cytoplasm
The Rev protein of the HIV virus normally shuttles back and forth between the nucleus and the cytoplasm to facilitate viral mRNA export. In the presence of the broad-spectrum antibiotic leptomycin B, Rev is found exclusively in the nucleus. Which statements indicate a possible way in which leptomycin B might alter the distribution of Rev?
a. Leptomycin B may bind to a component of the nuclear pore complex.
b. Leptomycin B could bind to Ran-GTP, facilitating its conversion to Ran-GDP.
c. Leptomycin B could bind to Ran-GDP to promote its conversion to Ran-GTP.
d. Leptomycin B could bind to the nuclear localization signal of Rev
e. Leptomycin B may inactivate the nuclear import receptor.
b. Leptomycin B could bind to Ran-GTP, facilitating its conversion to Ran-GDP.
You isolate a mutant of yeast and discover that the mutation is in the Ran-GAP. What will be the consequence of this mutation for protein transport?
a. Protein will be degraded
b. Protein will accumulate in the ER
c. Protein will accumulate in the nucleus
d. Protein will accumulate in the cytoplasm
d. protein will accumulate in the cytoplasm
To be imported into a mitochondrion, proteins must be __________.
a. Phosphorylated
b. Fully folded
c. Unfolded
d. Ubiquitinated
c. Unfolded
How do proteins with a nuclear export signal enter the nucleus?
a. These proteins are transported into the nucleus by Ran-GDP.
b. These proteins also have a nuclear localization sequence.
c. Nuclear export receptors take these proteins into the nucleus.
d. These proteins are transported into the nucleus by Ran-GTP.
b. These proteins also have a nuclear localization sequence.
Facilitated diffusion:
a. requires passage of substances through an ion channel.
b. requires coupling to an energy-releasing system.
c. enables substances to diffuse through a membrane by attaching to a facilitative transporter.
c. enables substances to diffuse through a membrane by attaching to a facilitative transporter.
As we discussed in class, Ran-Gap is required to disassemble the Importin-alpha+CAS+Ran-GTP complex in the cytoplasm. Why?
a. Cas cannot bind importin-alpha in the presence of Ran-GTP
b. Ran in the GDP-bound state cannot bind importin-alpha
c. None of the above
d. Ran in the GTP-bound state cannot bind importin-alpha
b. Ran in the GDP-bound state cannot bind importin-alpha
Based on our class discussion about nuclear localization, which statement is correct?
a. In the nucleus, Ran is always in the GTP-bound form, whereas in the cytoplasm, Ran is in the GDP-bound state
b. The Ran-GEF called RCC1 is only found in the cytoplasm
c. In the nucleus, Ran is always in the GDP-bound form, whereas in the cytoplasm, Ran is in the GTP-bound state
d. The Ran-GAP is only found in the nucleus
a. In the nucleus, Ran is always in the GTP-bound form, whereas in the cytoplasm, Ran is in the GDP-bound state
The yeast TRP1 gene encodes an enzyme that catalyzes the first step of tryptophan synthesis. TRP1 is a cytoplasmic protein. You are conducting an experiment in which you fuse the mitochondrial matrix signal sequence to the N-terminus of TRP1 (mitoMatrix-TRP1). Cells carrying mitoMatrix-TRP1 were grown in the absence of tryptophan. Most cells died, but some grew. Why did some cells grow in the absence of tryptophan?
a. Because of a mutation in the Tim22 gene
b. Because of a mutation in the Tim9 gene
c. Because of a mutation in the Oxa1 gene
d. Because of a mutation in the Tim23 gene
e. Because of a mutation in the TRP1 gene
d. Because of a mutation in the Tim23 gene
Which of the following would be true in a sec12 mutant?
a. Secreted proteins would be stuck in an abundance of COPII vesicles.
b. The ER would be enlarged.
c. There would be a normal number of ER to Golgi vesicles.
d. COP I vesicles would fail to bud off the donor membrane.
e. Protein translation of a secreted protein would remain arrested
b. The ER would be enlarged.
In a genetic screen in yeast, sec genes were isolated that encode proteins essential for protein secretion. The sec4 gene encodes a small GTPase called Rab. A mutation in the sec4 gene causes the mutant Rab protein to stay in the GDP-bound form. What would be the consequences for this mutant?
a. The mutant sec4 protein could no longer bind to its effector
b. The v-SNARE protein would directly interact with the mutant sec4 protein
c. The v-SNARE protein would directly interact with the Rab effector at the target membrane.
d. The Rab effector protein was no longer associated with the target membrane
e. The v-SNARE and t-SNARE proteins could no longer interact
a. The mutant sec4 protein could no longer bind to its effector
T/F: The signal peptide binds to a hydrophobic site on the ribosome causing a pause in protein synthesis, which resumes when SRP binds to the signal peptide.
False
You have placed a nuclear localization signal (NLS) near the C-terminus of a protein that had an N-terminal signal sequence for ER import. Where would this protein go?
a. Into the plasma membrane
b. Into the nucleus
c. Into the ER
d. Into the Golgi
c. Into the ER
What is the phenotype of a mutant of Sar1 that remains in the GTP state?
a. Secreted proteins accumulate in vesicles
b. KDEL proteins remain in the Golgi
c. Lysosomal proteins reach the lysosome
d. COP II proteins disassemble before budding has finished.
e. Sar1 is free in the cytosol
a. Secreted proteins accumulate in vesicles
If you add a nuclear localization signal to the C-terminus of a type I membrane protein, where would this protein go?
a. Outside of the cell
b. Into the cytoplasm
c. Into the nucleus
d. Into the ER
d. into the ER
BiP is usually a component of the ER lumen. You add the following hydrophobic amino acids to the middle of the BiP protein sequence: FVAILALWFMLIVGVFILVV. Where would you now predict the bulk of BiP to localize?
a. Secreted outside the cell
b. The ER lumen
c. The ER membrane
d. The lysosome
e. The plasma membrane
e. plasma membrane
Which of the following would be true in a cell mutant for a t-SNARE that normally localizes to the cis-Golgi?
a. The ER would be enlarged.
b. There would be a normal number of ER to Golgi vesicles.
c. Secreted proteins would be stuck in an abundance of COPII vesicles.
d. COP I vesicles would fail to bud off the donor membrane.
e. Protein translation of a secreted protein would remain arrested.
c. Secreted proteins would be stuck in an abundance of COPII vesicles.
What would be the outcome of a double mutant defective in both sec12 AND the cis-Golgi t-SNARE?
a. There would be a normal number of ER to Golgi vesicles.
b. The ER would be enlarged.
c. COP I vesicles would fail to bud off the donor membrane.
d. Secreted proteins would be stuck in an abundance of COPII vesicles.
e. Protein translation of a secreted protein would remain arrested.
b. the ER would be enlarged
What is the fate of a protein with no sorting signal?
a. None of the above
b. Retained in the ER
c. Remain in the cytosol
d. Retained on the translating ribosome
e. Routed to the nucleus
c. Remain in the cytosol
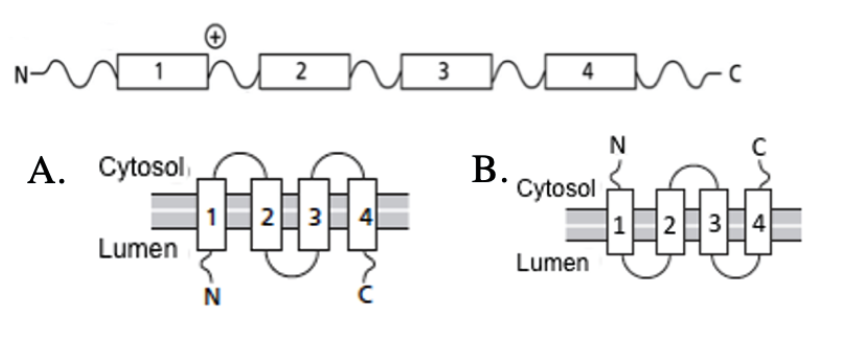
A schematic of a membrane protein is presented below. The boxes illustrate membrane-spanning segments. What will the topology of this protein be in the ER?
A
B
A
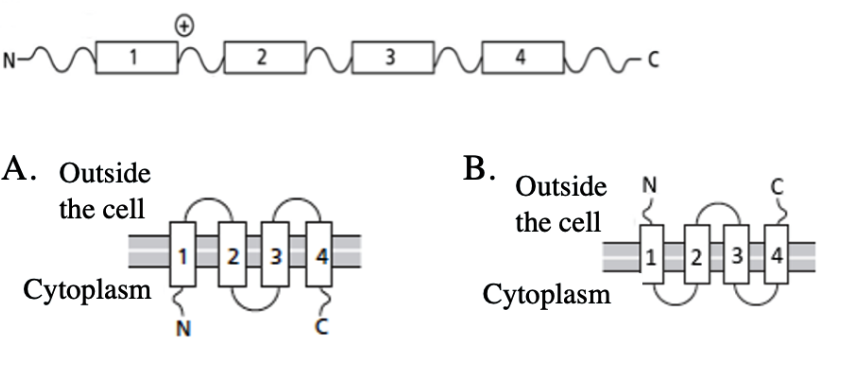
A schematic of a membrane protein is presented below. The boxes illustrate membrane-spanning segments. What will the topology of this protein be in the plasma membrane?
A
B
B
Arf1 is the G protein for COP I vesicles and acts analogously to Sar1 for COP II vesicles. Where would you expect to find the GEF for Arf1?
a. Cytoplasm
b. The cis-Golgi membrane
c. Associated with COP I components
d. The ER membrane
e. On COP II vesicles
b. The cis-Golgi membrane
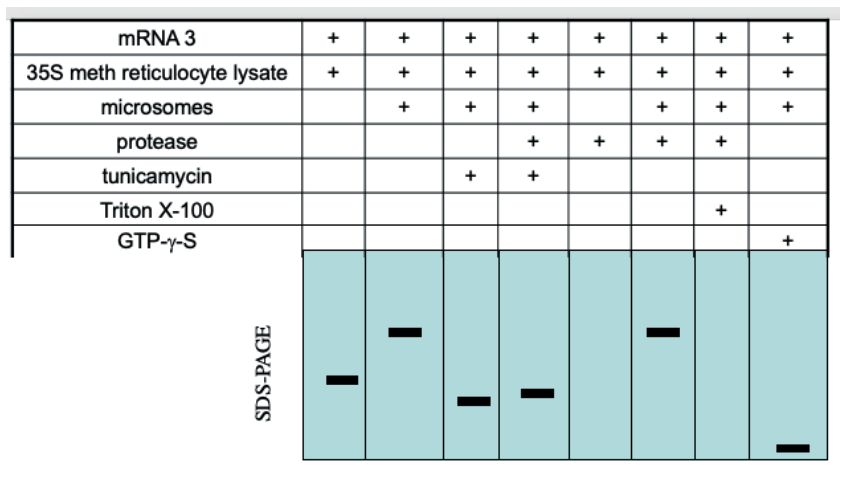
GTP-gamma-S is a non-hydrolyzable form of GTP. When GTPase binds to GTP-gamma-S, it becomes locked in the GTP conformation. Why is the band in the last lane much smaller?
a. SRP is unable to bind the SRP receptor
b. The translocon is not open
c. SRP is unable to release from the ER membrane
d. SRP is unable to bind the signal peptide
c, SRP is unable to release from the ER membrane