gene technology
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
85 Terms
what is the genome
all the genes / genetic material of an organism
what is the proteome
the full range of proteins that can be produced by an organism at a given time
what is larger, the genome or the proteome
the proteome
what are the benefits of developing a complete map of the human genome
develop a better understanding of which genes are responsible for certain genes
production of more effective medicines for genetic diseases and cancer
aids the development of vaccines
tracing evolutionary relationships
allows for genetic screening
suggest why it might be difficult to work out a mammals proteome from its genome
proteins are only produced when the genes for them are switched on
much of the genome consists of introns (non-coding DNA)
production of some proteins is influenced by multiple genes
scientists can gain information about antigens from sequencing the proteome of pathogens, why might this be useful
antigens are found on the surface of pathogens and pathogens can cause disease
knowledge about antigens may make it easier to identify pathogens
knowledge about the antigens found on pathogens may facilitate the production of effective vaccinations and medical treatments for particular diseases
suggest and explain how identifying the parasite that causes malarias’s genome can be used to speed up the development of a vaccine
the genes showing the largest level of variance are likely to be the antigens (because antigens vary to try and evade the host’s immune system)
the identification of these vary genes, allows scientists to specifically focus on this area of the genome, instead of wasting their time looking at areas of the genome that don’t affect antigen production
once the antigenic genes have been worked out, scientists can then synthesise the proteins produced by these genes and inject them into people (as vaccines) to determine whether it causes an immune response
why is it hard to determine the human proteome (eukaryotes)
eukaryotes have introns in the DNA (non coding sections of DNA)
eukaryotes have regulatory DNA where genes can be switched on by transcription factors to produce proteins- expression (on/off) of these genes is constantly changing)
why is the proteome of a prokaryotic pathogen easier to determine than the proteome of a human
the prokaryotic proteome is smaller than the human proteome
prokaryotes don’t have introns (non coding DNA), whereas eukaryotes have many introns incorporated into it
humans have more regulatory DNA than prokaryotes (DNA where genes can be switched on by transcription factors to produce proteins- expression (on/off) of these genes is constantly changing)
why is it necessary to sequence the DNA of several organisms of the same species to develop an accurate genetic template of the species
individuals show genetic variation, so may have differences in there genes and alleles
there may be mutations within the DNA sequence of an individual
using a larger sample size/more individuals provides a more accurate representation of the population
how can DNA sequencing be used to determine evolutionary relationships
species that are more closely related will have more similar DNA/RNA sequences and protein sequences
DNA for human genome sequencing projects uses blood sample, which cells are used and why
white blood cells
these are the only cells in the blood that have nuclei (platelets and RBC’s don’t)
what are the benefits of automated DNA sequencing methods compared to the original manual methods
algorithms are used to analyse data on genetic sequences- reduces the risk of errors in the sequencing process
scientists can make comparisons of gene sequences with known sequences stored in the data base
can make genome comparisons faster
computers can immediately identify new genes and alleles
what are the advantages of making scientific data public, prior to publication
any medical treatments developed by using the public data would be cheaper (don’t have to pay to use the data)
speed of the research process would be faster because other scientists can access the information for collaboration
what are the disadvantages of making scientific data public, prior to publification
have to rely on public funding- charity or government funding
the data could be used by other researchers for unethical and illegal research
what does DNA sequencing allow for
the base sequence of an organism to be identified and recorded
the genetic code is ‘universal’, what does this mean
almost every organism uses the same 4 nitrogenous bases (ATCG)
the same codons code for the same amino acids in all living things (genetic information is transferrable between species)
what is recombinant DNA (rDNA)
DNA that has been artificially changed by combining lengths of nucleotides from different sources (different species e.g. bacteria)
this process works because the genetic code is universal (same 4 bases and codons will code for the same amino acids)
Why are bacteria able to use human DNA to produce human proteins
The genetic code is universal (same for prokaryotes and eukaryotes)
The mechanism of transcription and translation are universal
what happens in genetic modification
fragments of DNA from one organism have to be transferred into another organism
this results in a genetically modified organism which will contain recombinant DNA (rDNA)
what are some applications of recombinant DNA technology
Production of recombinant proteins (such as insulin) from bacterial cells
Gene therapy- inserting healthy copies of mutated genes into human cells
Pest-resistant crops- better yield
Herbicide-resistant crops- better yield
More nutritious crops (with added nutrients)
increased growth hormone production in farmed animals- better yield
describe the process of in vivo gene cloning
The desired gene/DNA fragment is identified
The gene/DNA fragment is isolated and extracted (isolation)
The gene/DNA fragment is then inserted into a plasmid vector (insertion)
allow the bacteria to multiply (transformation)
The bacteria cells that have taken up the gene/DNA fragments are identified using genetic markers (identification)
The genetically modified cells are then cloned (growth/cloning)
describe the process of in vitro cloning
the desired gene / DNA fragment is identified
the gene / DNA fragment is isolated and extracted (isolation)
DNA fragments are amplified in the polymerase chain reaction (PCR)
how is the desired gene/DNA fragment isolated and extracted
the gene with the specific characteristic that is required can be obtained in three ways:
extracting a gene from the DNA of a donor using restriction endonuclease enzymes
using reverse transcriptase to synthesise a single strand of complimentary DNA from the mRNA of a donor organism
synthesising the gene artificially using the gene machine
what are restriction endonucleases
they are enzymes found in bacteria used as a defence mechanism against viruses- where they cut the viral genetic material into smaller pieces at specific nucleotide sequences within the molecule
the restriction endonucleases are named according to the bacteria they are sourced from
how can a gene be extracted from the DNA of a donor using restriction endonucleases
the restriction endonuclease enzyme will cut a DNA double strand at the same place at a particular recognition sequence- normally around 6 base pairs
this can form sticky ends where the sugar-phosphate backbone is cut in an uneven/staggered way
or it can form blunt ends where the sugar-phosphate backbone is cut straight across
why are sticky ends preferred over blunt ends
when the DNA is cut to produce “sticky ends", two complimentary ends are created
This means that only DNA sticky ends produced with the same restriction endonuclease can join together
With blunt ends, there is no specificity to the cut
this means that any strand can join with any other strand are the ends are not specific and complementary
how can reverse transcriptase (enzyme) be used to synthesise a single strand of complimentary DNA from the mRNA of a donor organism
Use the mRNA that was transcribed for the desired gene
Once isolated, the mRNA is combined with a reverse transcriptase enzyme and nucleotides to create a single strand of complementary DNA
add DNA primer that will bind to the mRNA strand which signal reverse transcriptase where to start synthesising the DNA
this forms a DNA strand complimentary to the RNA strand
where can reverse transcriptase enzymes be sourced from
retroviruses
why is isolating the desired gene using reverse transcriptase preferred over using restriction endonucleases
mRNA doesn’t contain introns (non-coding DNA)- introns are spliced
how can e gene with the specific characteristic that is required can be obtained via artificial synthesis using a “gene machine”
due to technological advancements, scientists are becoming more aware of the proteome (base sequences for our proteins)
this makes it possible to synthesise genes artificially
as we know which amino acids are required, scientists can use computers to generate the nucleotide base sequences for genes to produce the proteins that we want
short fragments of DNA are produced first which are joined to make longer sequences of nucleotides, which are then inserted into vectors (such as plasmids)
what happens after the gene/DNA fragment is isolated
promoter and terminator regions are added to the fragments of DNA to ensure replication
what two ways can DNA fragments be amplified
in vivo cloning- use of plasmids and bacterial cells
in vitro cloning- PCR
how are the DNA fragments inserted into plasmids (insertion)
restriction endonucleases are used to cut plasmids and produce sticky ends that are complimentary to the desired gene’s sticky ends
the isolated DNA fragment with the correct sticky ends, promoter region and terminator region are mixed together with DNA ligase
DNA ligase catalyses the condensation reaction where phosphodiester bonds are formed between the nucleotides on two separate DNA strands
some plasmids will take up the gene and form recombinant plasmid DNA
what happens in transformation in in-vivo cloning
the vector (recombinant plasmid DNA) is transferred into bacterial host cells
to do this the cell membrane of the host cell must be more permeable- which can be done by mixing the bacterial cells with calcium ions and a sudden increase in temperature
only a few bacterial cells actually take up the recombinant plasmid DNA
these bacterial cells can be identified by using markers
why are bacterial cells used in in-vivo cloning
bacterial cells are used because the can divide and reproduce rapidly via binary fission and they are relatively easy to culture
why do we need to identify transferred bacteria
not all of the host cells (bacteria) will take up the recombinant plasmid DNA
this can be due to:
the recombinant plasmid DNA doesn’t get inside the cell
the plasmid rejoined to itself before the DNA fragment could join
the DNA fragment sticks to itself, rather than to the plasmid
how are bacterial cells that have taken up the recombinant plasmid DNA identified (identification)
there are different methods:
replica plating
fluorescent markers
enzyme makers
how can replica plating be used to see if plasmids have taken up the DNA fragment
plasmids have genes for ampicillin and tetracycline antibiotic resistance
depending on which restriction endonuclease is used to cut stick ends, the gene for either ampicillin or tetracycline antibiotic resistance could be removed
BamHI restriction endonuclease removes tetracycline resistance but ampicillin resistance remains
method:
individual bacteria cells are plated onto the master plate, replicating to form colonies
a replica plate is cultured with ampicillin, leaving only cells that contain plasmids with ampicilin resistance
a second replica plate is cultured with tetracycline, leaving only cells with ampicillin and tetracyclie resistance, so these plasmids haven’t taken up the DNA fragment
the cells with ampicillin resistance, but no tetracycline resistance are the desired cells
what are some disadvantages of replica plating
creating colonies of bacteria → antibiotic resistance
slow process
wasteful process
how can fluorescent markers be used to identify plasmids that have taken up the DNA fragment
plasmids containing a green fluorescent protein (GFP) incorporated from a jelly fish can be used
a restriction endonuclease cleaves a site at the GFP region should be used
plasmids that take up the DNA fragment / gene will not fluoresce under UV light → they are the bacterial cells (called transformed bacteria) that will be cultured, so the desired gene is cloned
plasmids that fluoresce under UV light will be destroyed
how can enzyme markers be used to identify whether a plasmid has taken up the DNA fragment
another gene marker is the gene that produces the lactase enzyme
lactase breaks down lactose in X-Gal and one of those products is blue in colour
a restriction endonuclease that cleaves the lactase producing gene should be used
if the insertion is successful and transformed bacteria is produced, then when the bacterial cells are cultured with X-Gal, the substrate will not turn blue as lactase isn’t produced → these are the bacterial cells that will be cultured so the desired gene will be cloned
what are transferred bacterial cells
bacterial cells that have taken up the recombinant plasmid DNA
what happens in the growth/cloning stage of in vivo cloning
the transformed bacteria will have been identified
these are the bacteria that:
aren’t fluorescent under UV
when cultured with X-Gal, the substrate won’t be blue
have only either ampicillin or tetracycline resistance (not both)
these bacterial cells will then be cultured in a fermenter, every time the bacterial cell divides via binary fission then the desired gene will be cloned
these bacterial cells with then synthesise the protein coded for by the desired gene
what is the purpose of PCR
to produce large quantities of specific DNA fragments from small quantities of DNA
what piece of equipment is used in PCR to change the temperature and control the amount of time sent at each stage
thermocycler
which is an automated machine
what are primers
short sequence of single stranded DNA
they are complimentary to the start and end of the DNA being copied
they provide a starting point for DNA polymerase
what does each PCR reaction require
target DNA
primers
DNA polymerase- Taq DNA polymerase (attained from bacteria that live in hot springs because this enzyme has a much higher optimum temperature so it won’t denature at high temperatures (72.) in the thermocycler)
Free DNA nucleotides- construction of the DNA strands
buffer solution- provide the optimum pH for the reactions to occur in
thermocycler
describe the principles of the polymerase chain reaction (PCR) as an in vitro method to amplify DNA fragments.
denaturation (heat to 95°C)
annealing (cool to 50-65°C)
extension (heat to 72°C)
what happens in the denaturation stage of PCR
DNA sample is heated to 95°C which causes hydrogen bonds between complimentary base pairs to break
this results in the DNA separating into two single strands
what happens in the annealing stage of PCR
the temperature is lowered to 50-65°C, allowing the primer to bind (anneal) to the 3’ end of the target DNA
the temperature is cool enough for hydrogen bonds to reform so that the primer is held in place
primers prevent the two DNA strands from rejoining/re-annealing
the primer provides a starting point for DNA polymerase, as it can only add nucleotides to the existing strand
what happens in the extension stage of PCR
the temperature is increased to 72°C, which is the optimum temperature for Taq Polymerase
Taq polymerase adds free DNA nucleotides (A,T,C,G) and forms phosphodiester bonds between adjacent nucleotides → forms a new DNA polymer
why is it better to start PCR with mRNA rather than DNA
mRNA is shorter than DNA and more specific to the genes you want to amplify
introns in DNA are spliced/removed in mRNA
what are the advantages of PCR/ in vitro cloning
automated machine (thermocycler) which is more efficient
rapid- 100 billion copies of DNA can be made within hours
doesn't require living cells (such as bacterial cells)- quicker and less complicated techniques are needed compared to in vivo cloning
what are the disadvantages of in vitro cloning/ PCR
there are more errors in the DNA fragments produced compared to in vivo cloning
doesn’t synthesise products- unless DNA is incorporated into living cells
what are the advantages of in vivo cloning
there are less errors in the DNA fragments produced (with exceptions of mutations)
product synthesis- transformed bacteria can easily be cultured to produce the desired product e.g. insulin
what are the disadvantages of in vivo cloning
slower process (days/weeks to produce large quantities) than in vitro cloning which takes hours
uses living cells (bacterial cells) and vectors (plasmids)
what is gene therapy, and describe its use in medicine
gene therapy uses various methods to alter a persons genetic material
scientists are able to do this as they have a better understanding of the human genome and therefore, which genes may cause certain diseases
scientists can insert corrected copies of genes into patients to replace mutated genes to treat genetic diseases such as:
cystic fibrosis
severe combined immunodeficiency (SCID)
cancer
how does a normal functioning CFTR channel differ in function to a mutant CFTR channel (cystic fibrosis)
normal functioning CFTR channel- moves chloride ions out of the cell
mutant CFTR channel- doesn’t move chloride ions out of the cell → water potential in the lumen remains low → water doesn't move out of the cell via osmosis → mucus remains viscous and sticky
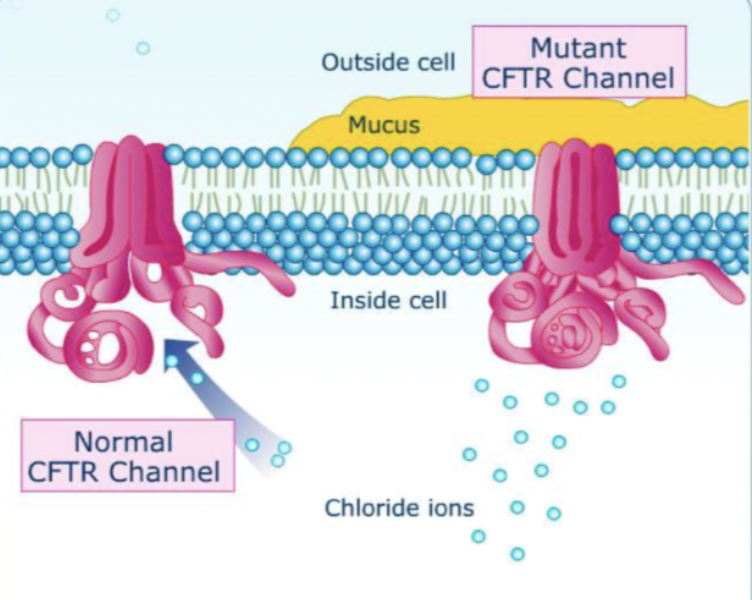
what is cystic fibrosis caused by
a recessive mutated allele
the mutation is a deletion mutation that removes AAA (lysine)
what are the effects of cystic fibrosis
thicker mucus in upper airways → increased risk of lung infection
thicker mucus on alveolar surface → greater diffusion distance → reduced rate of gas exchange and O2 intake
thick mucus can block pancreatic ducts → results in digestive enzymes not reaching the small intestine → poor digestion and absorption of nutrients
thick mucus can block sperm ducts → infertility
what are two ways that gene therapy can be used to treat cystic fibrosis
gene replacement: mutated allele is replaced by a functional healthy gene (lysine won’t be removed → there will be functional CFTR channels
gene supplementation: one or more healthy copies are inserted. The genes added have dominant alleles so the are expressed over the recessive mutated alleles
what are the two types of gene therapy
somatic cell gene therapy
germ-line gene therapy
what is somatic cell gene therapy
somatic gene therapy targets the non-reproducing (somatic) cells (effects only affect the individual, not any offspring they may have)
effects are temporary as these cells are commonly replaced or may die → treatment must be repeated every few days
method:
a functional gene is inserted into the patients somatic cells using vectors such as viruses to deliver the gene into target cells (e.g. lungs for cystic fibrosis gene therapy)
the inserted gene will now produce the correct protein and the symptoms of the disease will be reduced or eliminated
give some examples of somatic cells
red blood cells
epithelial cells
lung cells (alveolar cells)
nerve cells
liver cells
muscle cells
what are some social and ethical issues with somatic gene therapy
lots of injections required (somatic cells die and are placed) → expensive treatment → not accessible to all people
people might become less accepting of disabilities as they become less common
doping in sports- genes in muscle cells are altered to increase EPO hormone production which promotes the formation of RBC’s
what is germ line gene therapy
it involves inserting functional genes into early embryos
the genetic change is permanent and heritable for both the individual and any offspring they may come to have themselves
it eliminates the faulty gene that may cause certain diseases
what are the ethical issues with germ line gene therapy and why is somatic cell gene therapy more ethical
can’t obtain consent from future individuals affected by the therapy (future individuals aren’t affected in somatic cell gene therapy as the changes aren’t heritable)
unknown long term effects can be passed onto descendants (changes aren’t heritable and passed on in somatic cell gene therapy
potential for misuse- such as designer babies (somatic cell gene therapy is generally limited to only the treatment of disease)
how can a virus vector be used to treat cystic fibrosis
uses an adenovirus (which is harmless) that is known to cause respiratory infections by injecting DNA into epithelial cells in the lungs
the adenovirus is modified so it can’t replicate
the adenovirus is cultured in epithelial cells alongside modified plasmids that contain the normal CFTR gene inserted
the CFTR gene in the recombinant plasmid DNA is taken up by some of the adenoviruses
the adenoviruses that have taken up the recombinant plasmid DNA are identified using markers and extracted from the epithelial cells
the modified virus is then introduced to the patients nostrils to hopefully produce the correct CFTR protein when they ‘infect’ the patient’s lung’s epithelial cells
this works because the virus will inject it’s own DNA containing the normal CFTR gene into the DNA of the epithelial cell, so the gene will be transcribed and translated to produce a normal functioning CFTR protein → thick and viscous mucus won’t build up as chloride ions are transported out of the cell normally
what is an issue with using viral vectors in somatic cell gene therapy
Viruses may mutate and cause infections
They might trigger an immune response so the virus is destroyed before being effective for treatment
Gene may be inserted into the genome that leads to a harmful genetic mutation- insertion mutation that leads to a different amino acid being encoded for
how can a plasmid vector be used to treat cystic fibrosis
uses in vivo cloning to produce transformed bacteria which contain recombinant plasmid DNA with the function gene
the plasmids are removed and wrapped in lipid molecules to produce a liposome
the liposome is administered to the patient’s nostrils using a nasal spray
the liposome moves into the patients epithelial cells in the lungs through the phospholipid membrane (because liposomes are lipid soluble), delivering the plasmid with the functional CFTR gene which will then be expressed → thick and viscous mucus won’t be produced as chloride ions are transported out of the cell normally
what is severe combined immune deficiency (SCID)
an inherited disorder
inability to produce mature T cells → can’t carry out the cellular response
inability to produce mature B cells → can’t produce antibodies involved in the humoral response
what are labelled DNA probes and DNA hybridisation used for
identifying and locating a particular length of DNA
to treat genetic diseases, the faulty gene that needs to be treated must be located
what are DNA probes
A DNA probe is a short length of single stranded DNA that has a known base sequence that is complimentary to the base sequence of a ‘known faulty/harmful allele’
the probe is attached to a radioactive or fluorescent label that indicates its position
how are DNA probes created
the DNA probe can be artificially synthesised using a gene machine (because we know the base sequence of the harmful allele)
how is single stranded DNA on the nylon membrane created, so that DNA probes can anneal
a cell sample is taken from a patient → can gather cheek cells using a swab inside of the patients mouth
The DNA is extracted from the cell sample and purified
the DNA sample is amplified using PCR
restriction endonucleases are used to shorten the DNA molecules (because the whole DNA molecules are too long to be analysed in one go)
the smaller DNA fragments are separated using gel electrophoresis
the negatively charged DNA fragments move through the pores in the gel towards the positively charged electrode
smaller DNA fragments are able to move at a faster rate through the pores, so they travel further
the DNA fragments separate according to size and charge producing bands in the gel
the bands of gel are transferred to a nylon membrane
the DNA fragments on the nylon membrane are single stranded- this is done by breaking the hydrogen bonds between complimentary base pairs
how are DNA probes and DNA hybridisation used together to indicate whether a specific harmful allele is present in a DNA sample
DNA probes are added to the nylon membrane
DNA probes have a specific base sequence that is complimentary to the harmful allele
the DNA on the nylon membrane is single stranded so the DNA probe can anneal to any complimentary strands
if the harmful allele is present in the DNA sample, then hydrogen bonds will form between the complimentary base pairs of the DNA strand and the DNA probe
before the DNA probes are identified the nylon membrane is washed to remove any excess DNA probes that haven’t formed hydrogen bonds (prevents false positive)
DNA probes with fluorescent label- UV light is used to detect their position
DNA probes with radioactive label- X-rays are used to detect their position
if the label shows up on the DNA fragments then the DNA contains the harmful allele
what is genetic screening used for
to see if you have alleles present in your genome for a particular genetic disorder
what is the process of genetic screening
Person’s DNA
a sample of DNA is taken from the person
the DNA sample is amplified using PCR
restriction endonucleases are used to shorten DNA molecule into smaller DNA fragments
DNA fragments are separated using gel electrophoresis → produces bands in the gel
the bands are transferred to a nylon membrane
the DNA fragments on the nylon membrane are single stranded- hydrogen bonds between complimentary bases are broken to form single strand from double strand
DNA probes
DNA probes are synthesised from a gene machine and have a complimentary base sequence to the allele causing the genetic disorder the person is being screened for
DNA probes are radioactively labelled
if the harmful allele is present in the single strand of DNA then the DNA probe will form hydrogen bonds with the complimentary bases and anneal to the strand
nylon membrane is washed to removed excess DNA probes that didn’t anneal
X ray films /autoradiography can then be used to detect radioactively labelled DNA probes that have attached to the strand of DNA → to tell whether the harmful allele is present in the persons DNA
what are the advantages of genetic screening
people can plan ahead about reproduction → might not want to pass on allele for disease to their children → can adopt instead
individuals can make lifestyle changes to reduce chances of developing the diseases (breast cancer)
these people can participate in research and clinical trials → help develop understanding and treatments of genetic diseases
what are some disadvantages of genetic screening
if the disease is incurable then it might cause the person anxiety and/or depression
the person might not actually develop the disease and would’ve still suffered from anxiety from thinking they might develop it
it could increase the price of someones life insurance- which they may not be able to afford
Why might allele's for diseases that develop later on in life be passed on in human populations
alleles are passed on through reproduction
symptoms might develop late in an individual → don’t know they have it before they reproduce and pass on the allele
describe the role of genetic counselling
used to help people understand and process their genetic screening results
they can also inform individuals of possible results before they have a screening
what may genetic counsellors discuss with a patient
the chances of the individual developing an inherited disease
the chance of an individual having a child that develops the genetic disease
the lifestyle changes that can be made to reduce the risk of developing the disease
possible treatments for the patients if they do develop the disease