Chapter 4 - 3D Structures of Proteins
1/51
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
52 Terms
conformation
3D shape and arrangement of atoms that depends on the rotation of bond(s)
native conformation
each protein folds into a single stable shape
biological function of proteins are dependent on
proteomics
study of large sets of proteins like the entire complement of proteins produced by a cell
globular proteins
usually water soluble, compact, roughly spherical, tightly folded
hydrophobic interior, hydrophilic surface
ex: enzymes, carriers and regulatory proteins
fibrous proteins
provide mechanical support
components of large subcellular or extracellular structures
ribosomes, cilia
assembles into large cables or threads
ex: alpha keratins (hair and nails), collagen (tendon, skin, bones, teeth)
primary structure
structure of polypeptide or protein resulting from amino acid linear sequence in polypeptide chain

secondary structure
structure of polypeptide or protein resulting from H-bond interactions between peptide groups relatively close to each other
can be on same or different polypeptide chain
major structures: alpha helices & beta strands (and sheets)
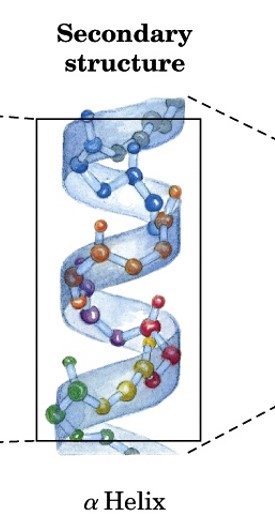
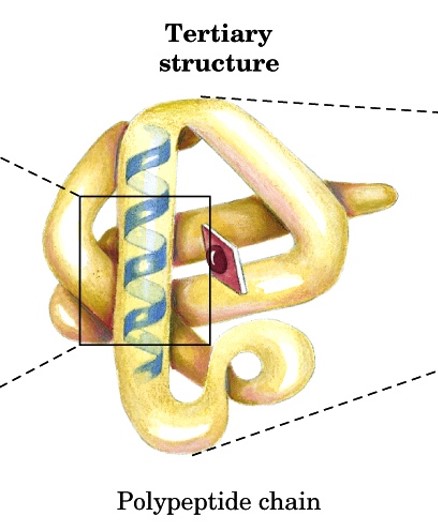
tertiary structure
structure of fully folded polypeptide chain into closely packed 3D structures
aka supersecondary structures
includes all AA residues (side chains)
stabilized by noncovalent interactions between side chains
motids in supersecondary structures
reoccurring protein structures
helix-loop-helix
coiled-coil
helix bundle
βαβ unit
hairpin
β meander
greek key
β sandwich
helix-loop-helix
two helices connected by a turn

coiled-coil
two amphipathic α helices interact in parallel through hydrophobic edges

helix bundle
several α helices that associate in antiparallel manner to form bundle

βαβ unit
two parallel β strands linked to intervening α helix by two loops

hairpin
two adjacent antiparallel β strands connected by β turn
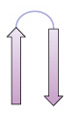
β meander
antiparallel sheet composed of sequential β strands connected by loops or turns
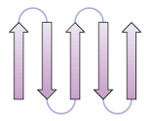
greek key
4 antiparallel strands
strands 1, 2 in middle
strands 3, 4 on outer edge

β sandwich
stacked β strands or sheets
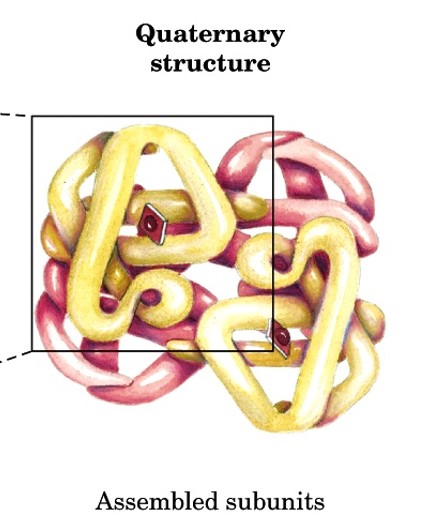
quaternary structure
arrangement of 2+ polypeptide chains into a multiunit molecules
subunits have defined stoichiometry and arrangement that are held together by many weak noncovalent interactions
X-ray crystallography
used to determine 3D conformation of proteins
beam of x-rays are aimed at crystal protein molecule
catalytic activity of enzymes in crystalline state shows proteins crystallize in their vivo native conformation
extra: Dorothy Crowfoot Hodgkin received Nobel prize in 1964 for determining structure of VitB12 using this technique

nuclear magnetic resonance (NMR)
used to determined structure of larger macromolecules like carbs, nucleic acids, and small-average proteins in solutions
shows conformational changes, protein folding, disulfide bridges, and interactions with other molecules
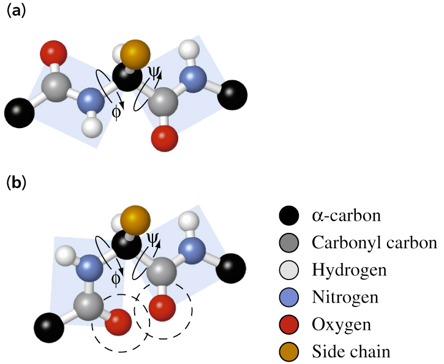
conformation of peptide groups
consist of 6 atoms
some have double bonds that restrict their conformation to trans or cis
cis is less favorable due to steric interference of alpha carbon side chains
trans is more favorable because two alpha carbons are on opposite sides and corners of the planar peptide group
has repeating N-Cα-C backbone
rotation about N-Cα (ϕ, phi) and Cα-C (ψ, psi) is possible
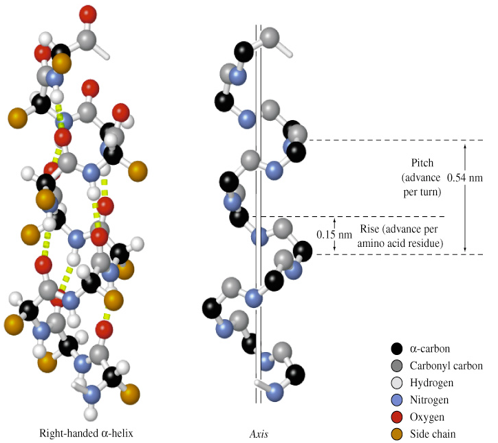
properties of α-helix
C=O forms H-bond with amide H of residue n+4
all C=O point to C-terminus (helix has dipole with (+) N, (-) C-termini)
stabilized by many H-bonds (parallel to long axis of helix)
Pitch = 0.54 nm
Rise = 0.15 nm along long axis
3.6 AA per turn
most are right-handed (from C-terminus, going down, helix turns clockwise)
extra: Linus Pauling & Robert Corey won Nobel prize in 1954)
β strands
polypeptide chains that are almost fully extended
stabilized by H-bonds between C=O and -NH on adjacent strands
β sheets
multiple β strands arranged side-by-side
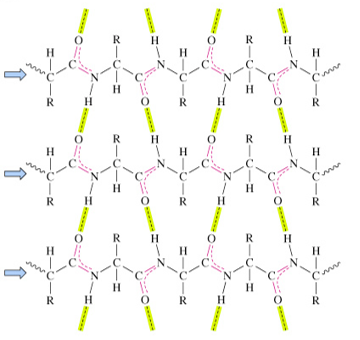
parallel β sheets
strands that run in the same N- to C-terminal direction
distorted H-bonds
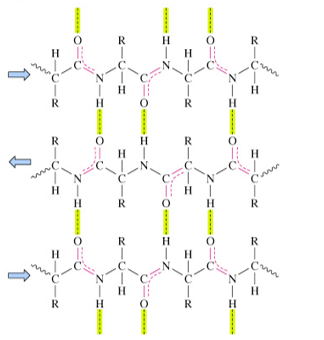
antiparallel β sheets
strands run in opposite N- to C-terminal directions
H-bonds are nearly perpendicular to chains
more stable than parallel chains
interactions of β sheets
project alternately above and below plan of β strands
one surface may consist of hydrophobic side chains that interact with hydrophobic residues
hydrophobic faces of β sheets can interact with hydrophobic side chains of amphipathic a-helices
how are helices and strands connected
loops and turns allow peptide chain to fold back on itself to make compact structure
loops
often contain hydrophilic residues
found on protein surfaces
turns
loops containing 5 or less residues
beta turns (reverse turns)
connect different antiparallel beta strands
domains
independently folded, compact units in proteins
size: ~25 - ~300 AA residues
connected by loops, bound by weak interactions between side chains
image contains 3 domains
all α domain
domain that consists of only α helices and loops
all β domain
domain only consists of β sheets and non-repetitive structures that link β strands
mixed α / β domain
domain containing supersecondary structures like αβα motif, regions of α helix and β strands alternate
α + β domain
domain consists of local clusters of α helices and β sheets in separate, continuous regions of polypeptide chain
domain function
binding small molecules
catalyzing single reactions
domain structure
interfaces provide crevices, grooves, and pockets on surface of protein for binding or catalytic sites
denaturation
disruption of native conformation of a protein
loss of biological activity
minimal energy required
caused by heat or chemicals (chaotropic agents and detergents)
some can be refolded/renatured
how do chemicals denature proteins
high conc. of chaotropic agents (urea, guanidinium salts, SDS) allow water to solvate nonpolar groups inside proteins
2-mercaptoethanol reduce disulfide bonds
folds
combo of secondary structures that form core of domain
folded proteins occupy low energy well = more stable
proteins fold spontaneously and rapidly (<1 sec)

hydrophobic effect
nonpolar side chains associate with each other causing polypeptide chain to collapse to molten globule
driving force is large increase in entropy from water released
hydrophobic collapse occurs at same time as formation of secondary structures
molecular chaperones
increase rate of correct folding
prevents incorrect formation of folded intermediates
can bind to unassembled protein subunits
most are heat shock proteins (synthesized as temp increases)
hydrolysis of ATP is needed

collagen
major protein in connective tissue (25-35% of total protein in mammals)
tendons (ropelike fibers) and skin (loosely woven fibers)
consists of 3 left-handed helical chains coiled around each other in right-handed supercoil
3 AA per turn
rise: 0.31nm per residue
more extended than α helix
globin
protein component of Mb and Hb
His-93 is complexed to iron atom
His-64 forms H-bond with oxygen
myoglobin (Mb)
monomeric protein that facilitates the diffusion of oxygen
composed of 8 α helices
interior all hydrophobic AA
extra: John Kendrew determined the structure of myoglobin
hemoglobin (Hb)
tetrameric protein that carries oxygen in the blood
α2β2 tetramer (2 α globin, 2 β globin subunits)
each globin similar in structure to myoglobin and has heme group
extra: Max Perutz determined the structure of hemoglobin
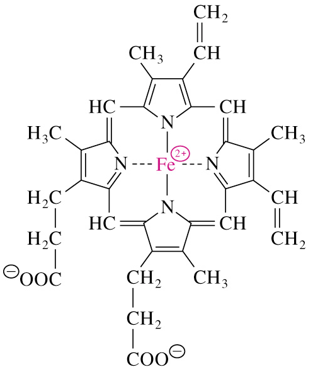
heme
consists of a tetrapyrrole ring called protoporphyrin IX complexed with iron
heme of Mb and Hb binds oxygen for transport
occupies hydrophobic cleft formed by 3 α helices and 2 loops
oxymyoglobin
oxygen bearing myoglobin
6 ligands are coordinated to ferrous ions in octahedral symmetry
oxygen is coordinated between iron and imidazole side-chain of His-64
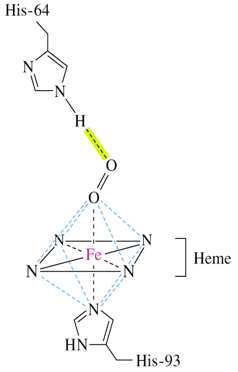
deoxymyoglobin
oxygen-free myoglobin
allosteric interactions
ex: regulates oxygen binding and releasing from Hb
allosteric effectors (modulators)
bind to protein at site separate from functional binding site
regulated activity of allosteric protein