Week 3 - Genomes and Chromosomes
1/79
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
80 Terms
What do genomes entail?
All the genetic material in an organism.
What genetic material is found in all living things?
DNA.
What genetic material is found in non-living things (ie. viruses)?
Sometimes DNA and sometimes RNA.
Is saying nucleotide pairs long and base pairs long the same thing?
Yes.
Can genomes vary in size?
Yes.
How many base pairs are in one human genome?
3 billion.
What is the genome size of T4 Bacteriophage?
169,000 base pairs long.
What is a model bacterial organism?
E. Coli.
Previously, we mentioned that Mitochondria and Chloroplasts originated from prokaryotes. If the model bacterial organism, E. Coli, is 4.6 million base pairs long, but human mitochondria and chloroplasts are only about 16,000 and 121,000 base pairs long now, respectively. What happened to all that DNA?
Some mitochondrial DNA has gotten into nuclear DNA.
If one human genome has 3 billion base pairs, how many base pairs are in each standard cell?
Since you get 1 genome from your mom and 1 genome from your dad, you have 2 sets of genomes)
Therefore, you get 23 chromosomes from your mom and 23 chromosomes from your dad. In total, each human then has 46 chromosomes, which is equivalent to 6 billion base pairs in each standard cell.
Is genome size always correlated to size of the organism or complexity of the organism?
No.
For example, a Newt is significantly smaller than a human and much less complex, and yet its genome is considerably larger to a humans.
What is the human genome made of?
50% is made of repetitive DNA
less than 1% of the human genome encodes for proteins
What is found inside unique sequences?
Introns
Exons
Non repetitive DNA
What happens to introns?
They are spliced out once RNA is transcribed.
What do exons do?
They encode for proteins.
What do non-repetitive DNA sequences, that are not introns or exons, do?
These sequences help determine which RNA sequences get transcribed in which cells and by how much.
What is within repeated sequences in the human genome?
Simple repeats
Segment duplications
Mobile Genetic Elements
What are simple repeats?
They are short DNA sequences, like codons, that are repeated multiple times (ie. CAGCAGCAG).
What are segment duplications?
Thousands to hundreds of thousands of base pairs that are duplicated.
What are mobile genetic elements?
They sometimes make a copy of themselves, paste themselves into the genome or cut themselves out of the genome.
What is found within mobile genetic elements?
Retrotransposons
DNA-only Transposon
LINEs
SINEs
What are DNA-only transposons?
They are just DNA.
What are retrotransposons?
They get made into RNA.
What are LINEs?
LINEs stand for Long Interspersed Nuclear Elements (≥500 base pairs). They encode proteins that help them move around the genome.
What are SINEs?
SINEs stand for Short Interspersed Nuclear Elements (<500 base pairs). They are short, repetitive DNA sequences that are found in eukaryotic genomes (they are non-coding).
Why do we package DNA?
This is because, when a genome is not packaged, even a small prokaryotic genome would occupy a large portion of the cells volume.
How is DNA condensed in prokaryotes?
Through folding and twisting around a protein.
What does condensed DNA in prokaryotes make?
Prokaryotic nucleoid.
What does the prokaryotic nucleoid do?
It organises DNA in prokaryotes.
Why is DNA so difficult to package?
It is VERY long and so packaging it into a nucleus, where it is still organised and usable, is quite a struggle.
Does a eukaryotic cell have 1 or 2 membranes?
It has 2 membranes:
inner membrane
outer membrane?
What is Chromosome Painting Hybridisation?
It is a technique where you paint chromosomes different colours?
Is PCR used in Fluorescence In Situ Hybridisation (FISH)?
Yes.
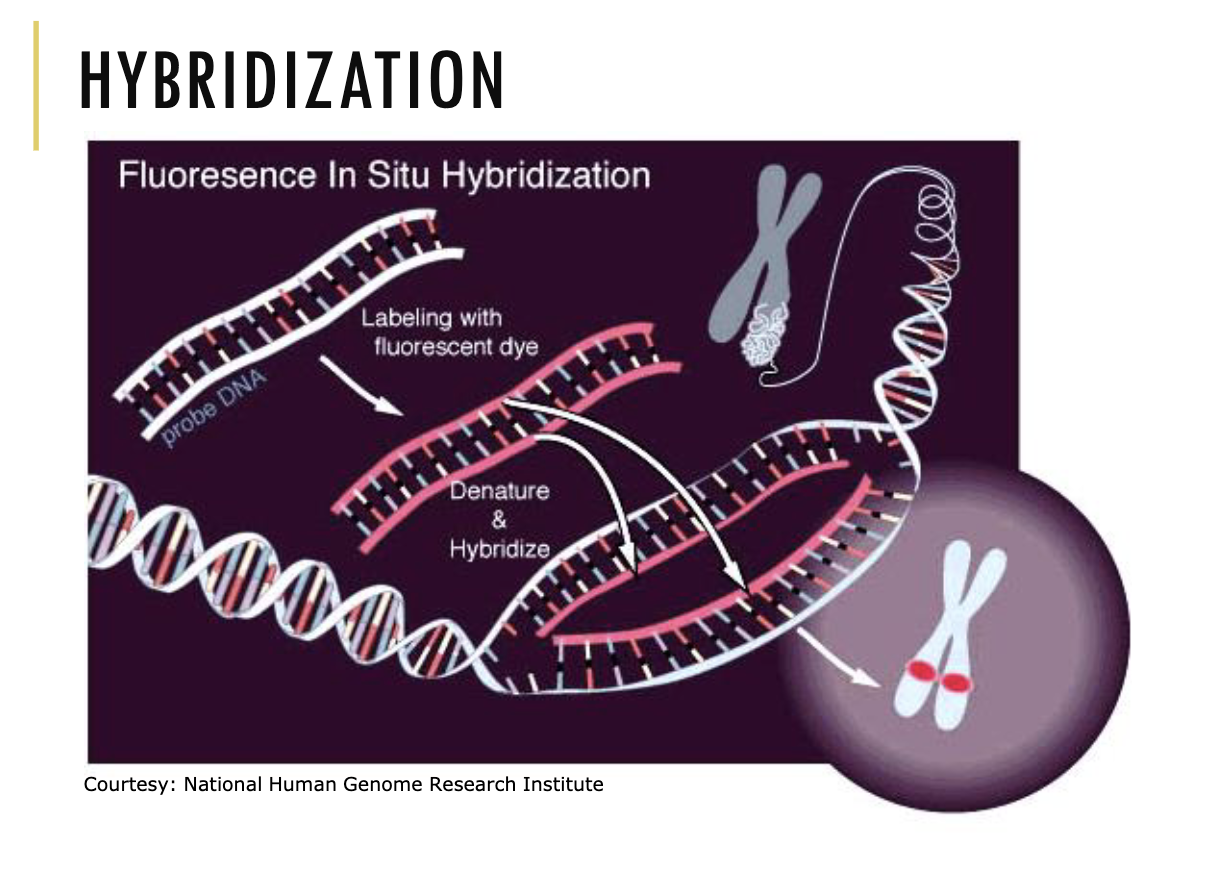
What is Fluorescence In Situ Hybridisation (FISH)?
It is a diagnostic technique to detect the presence of a specific DNA sequence.
How do we conduct Fluorescence In Situ Hybridisation (FISH)?
Collect probe DNA and mix it with a sample of DNA
Label the probe DNA with fluorescent dye so it shines a certain colour in the sample
Denature the probe DNA by heating it
Cool the probe DNA
If the DNA sequence you’re looking for is present in the sample, it will bind anti-parallel to complementary base pairs on the DNA sequence trying to be found and make that DNA sequence fluorescent
*if the DNA sequence is not there though, the probe will get washed away and you will see no fluorescence
What is probe DNA?
A single-stranded sequence of DNA or RNA that is used to identify specific sequences of DNA or RNA.
In FISH, when the sample is denatured and cooled, couldn’t the probe DNA and target DNA bind back to their own strands? How do we fix this?
Yes, they could bind back to their own strands.
To fix this:
A LOT of probe is put into the sample. Hence, making the probability that the probe binds to the target DNA higher.
Also, the procedure is carried out multiple times to double check that the target DNA is actually there.
Where do we get the probe DNA and fluorescent dye?
Chemists make them— well now robots make it because it’s THAT easy make.
How does chromosome painting hybridisation work?
It is the same procedure as FISH.
However, multiple probes are put into one sample to each target different DNA sequences. Also, each probe is labelled a different colour depending on the DNA sequence they’re looking for.
What is a karyotype?
It is a way to organise chromosomes based on their length and you typically put the sex chromosomes at the end.
What does each chromosome contain?
Each contain a single, long, linear DNA molecule and associated protein called Chromatin.
What are some characteristics of Chromatin?
They are a mixture of DNA and proteins which make up chromosomes found in our cells
They are tightly packaged but are still accessible for transcription, replication and repair
They are dynamic, meaning they can be condensed and de-condensed
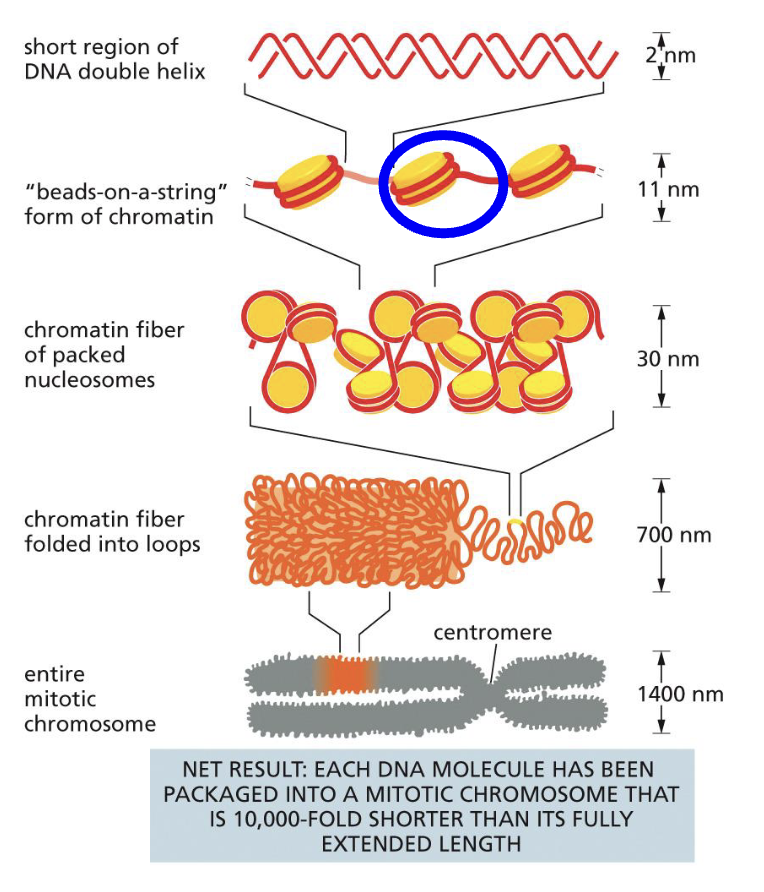
What are the levels of organisation for chromatin?
DNA double helix wraps around proteins 1 and 2/3 times. This is called ‘beads on a string’ form of chromatin
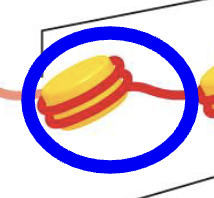
When looking at the individual protein wrapped with DNA, that is called a Nucleosome (the blue circle)
Nucleosomes can pack together to make a 30nm fibre
The fibre can then package into loops and make an entire chromosome
Is all chromatin 30nm?
NO, not all.
How long is linker DNA in the beads on a string form of chromatin?
80 base pairs long.
How long is the DNA wrapped around the protein in beads on a string form of chromatin?
147 base pairs long.
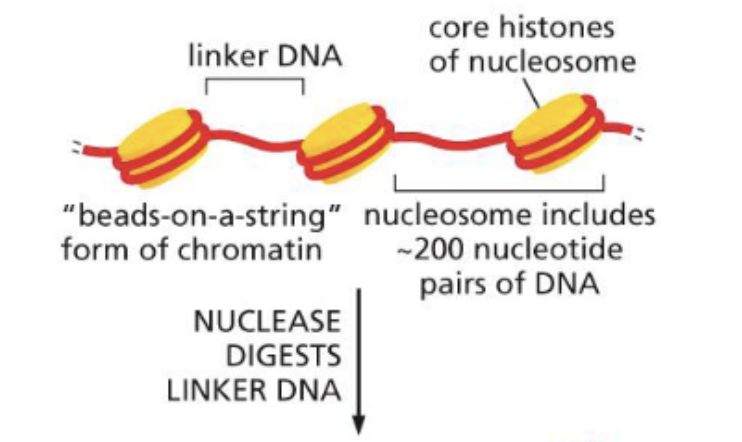
How do you open up a Nucleosome?
Use a nuclease to digest the linker DNA and break up the beads on a string form of chromatin
This then releases the nucleosome core particle (aka. the individual protein with the DNA wrapped around it)
To unravel the DNA from the protein, a high concentration of salt is used to dissociate them
One dissociated, you will notice that they separate into a histone octamer (mean 8 proteins) and a 147 base pair DNA double helix
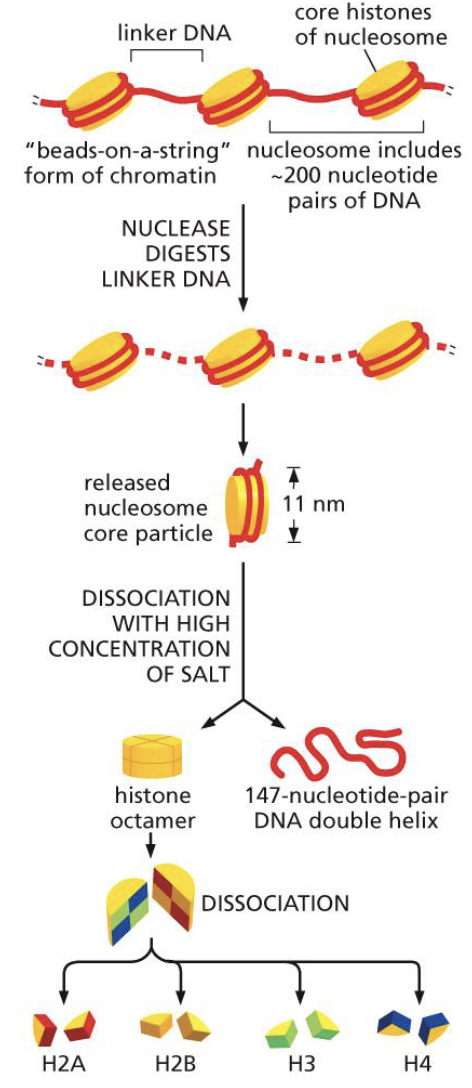
What are the characteristics of histone proteins?
Within the histone octamer (8 proteins), there are:
2 proteins of H2A
2 proteins of H2B
2 proteins of H3
2 proteins of H4
There is one linker histone, H1, which acts as a paper clip holding the DNA onto the histone (see picture)
Histones are very small proteins
Histones are rich in Lysine and Arginine
Histones have a positive charge that neutralises the negative charge on DNA
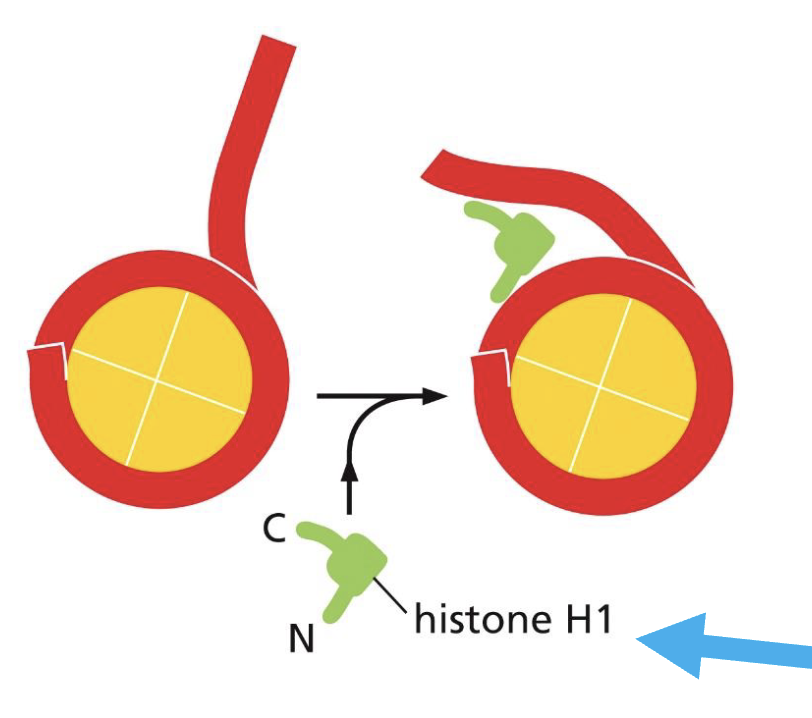
Can DNA still stick to the histone protein without the linker histone?
No, it will not be enough to stick. Hence, the linker histone is essential.
What does the nucleosome core particle refer to?
It refers to only the histone protein and the DNA wrapped around it (see image).
It DOES NOT refer to the linker DNA or H1 protein.
What does the nucleosome refer to?
It refers to the nucleosome core particle, the H1 protein and the linker DNA.
*the blue circle shows what is included
What allows the chromatin fibres to form loops?
Sequence-specific clamp proteins and cohesins are involved in forming chromatin loops.
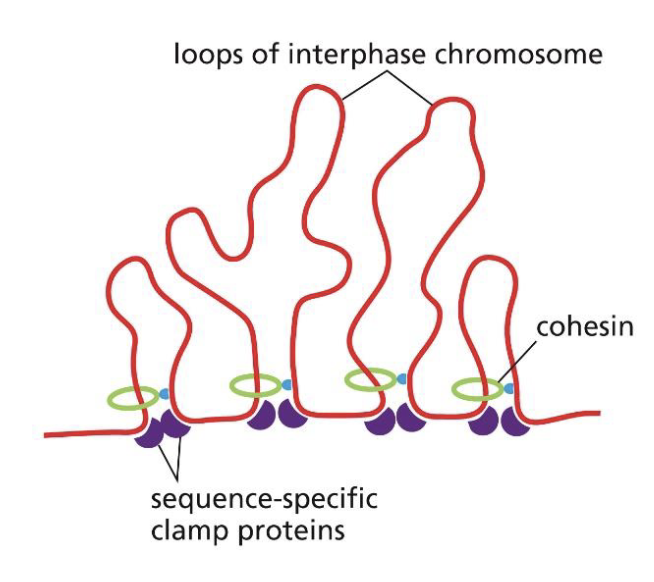
What allows chromatin to form loops that eventually make it become a chromsome?
As cells enter mitosis, condensins replace most cohesins to form double loops of chromatin. This then makes a compact chromosome.
Does packing and unpacking of DNA into a chromosome require ATP?
Yes.
What was the point of understanding the different levels of chromatin organisation?
As mentioned earlier in the lecture, since the DNA is so long, we need to find a way to compact it while making sure it’s still accessible. Hence, DNA is packaged into chromosomes by condensing chromatin, making their length 10,000 times shorter. That is much easier to deal with than its original length.
What is an example of packing and unpacking of DNA (chromatin) requiring ATP?
Chromatin Remodelling Complexes and Histone Modifying Enzymes
These are proteins that can make changes to chromatin structure and thus alter access to DNA for replication or transcription.
This requires ATP as they loosen the DNA's association with histones, making the DNA more accessible for replication and transcription.
What is another example of the packing and unpacking of DNA (chromatin) requiring ATP?
Heterochromatin and Euchromatin.
Define heterochromatin.
Highly condensed chromatin (≥30nm fibre).
Due to it being so condensed, the DNA is less accessible and gene expression is suppressed.
Define euchromatin
Relatively non-condensed chromatin (<30nm fibre) where genes tend to be expressed.
What region on the chromosome is heterochromatin?
Meiotic and mitotic chromatin chromosomes
the chromosomes are being moved around a lot so you want to make sure nothing gets unravelled
Centromeres
it’s the part that holds the sister chromatids together so you want it to be condensed
Telomeres
you don’t want your DNA double helix to be damaged so you want the ends to be condensed
Can the time heterochromatin spends being condensed vary?
Yes, there’s 2 ways:
It can always be condensed, in which case it would be referred to as constitutive
It can also be temporarily condensed. It would be called facultative at that point
Can the degree of condensation of Euchromatin vary?
Yes.
Euchromatin can be condensed in 2 ways:
quiescently
actively
What is quiescent euchromatin?
When the chromatin is not being transcribed or replicated.
What can change the degree of chromatin condensation (aka. change heterochromatin into euchromatin and vice versa)?
Covalent modification of histones
Chromatin remodelling complexes
RNA polymerase transcription complexes
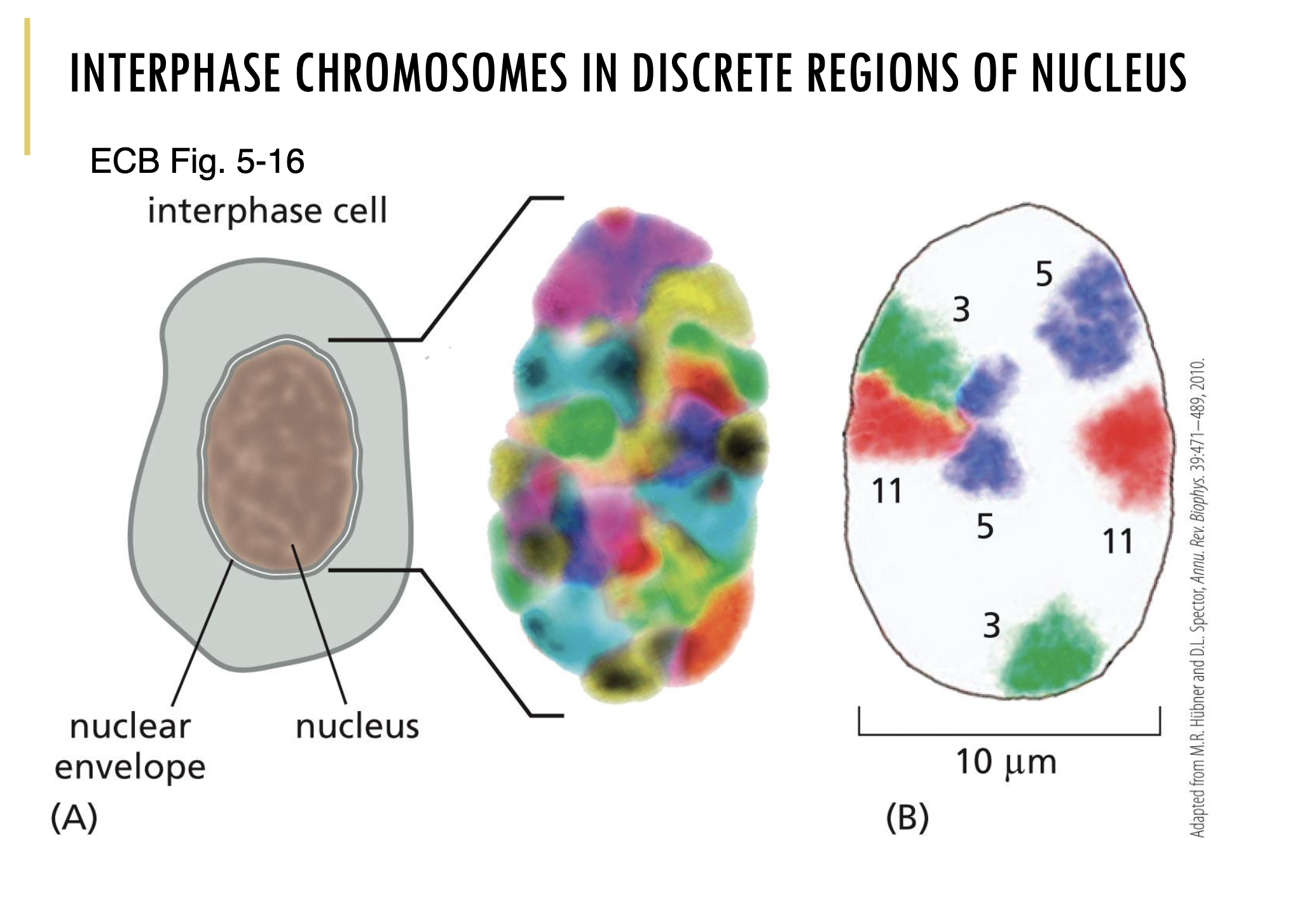
During interphase, are the chromosomes organised inside the nucleus?
Yes, interphase chromosomes are in discrete regions of the nucleus.
As you can see in the image, chromosome painting hybridisation was used to colour each chromosome type.
Through that, you can clearly see that each chromosome is organised into discrete regions in the nucleus during interphase.
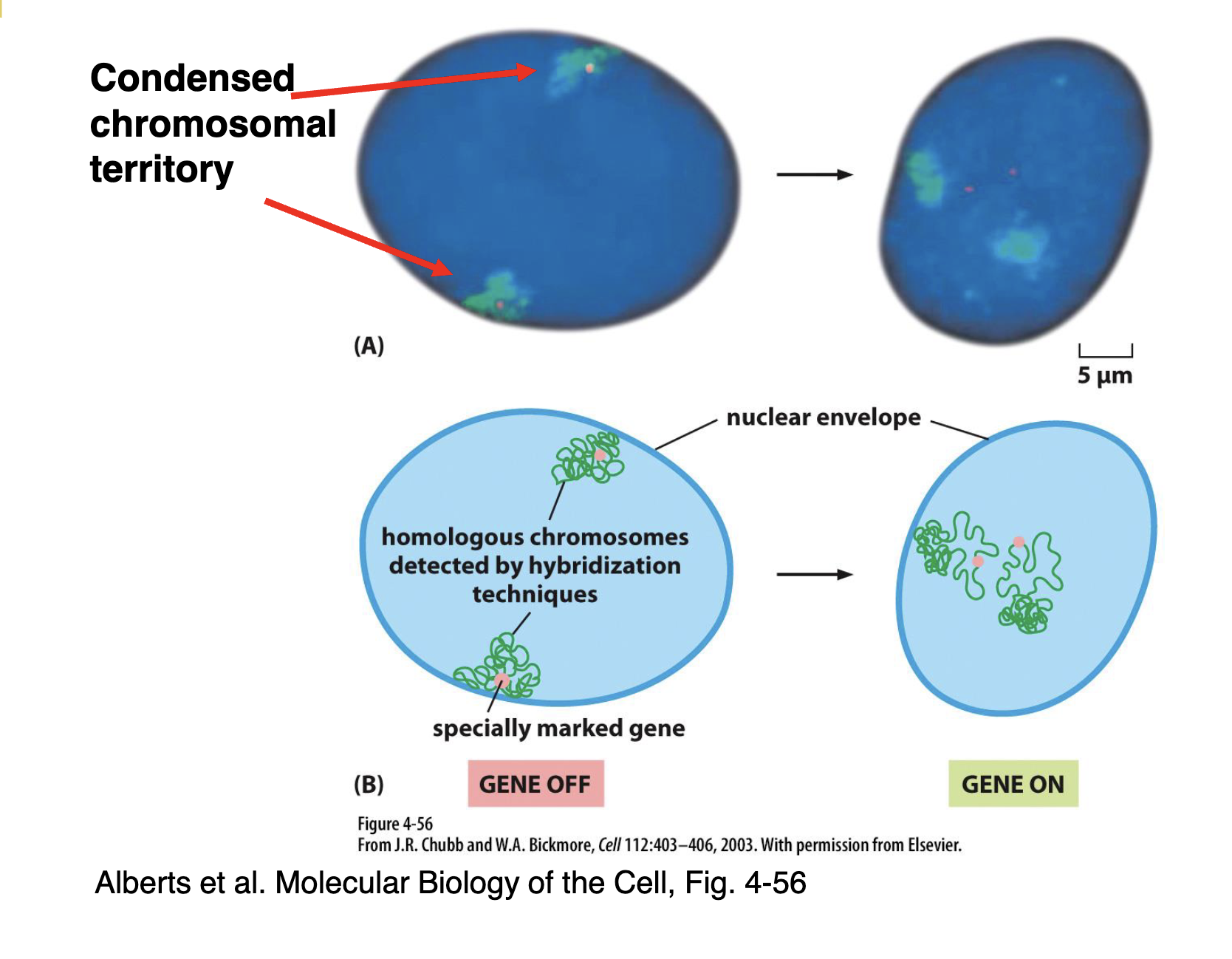
What happens to the orientation of genes within chromatin when they go from being suppressed to expressed?
Expressed genes are re-oriented (aka. de-condensed) within chromatin in order to be able to be read by RNA polymerase for transcription.
For example:
In the image, you can see homologous chromosomes (the chromatin is likely detected via chromosome painting hybridisation — shown in green and the gene is likely found by FISH — shown in peach) on opposite sides of the nucleus.
Also, the chromatin is very condensed and so the RNA polymerase cannot access the gene that’s ‘buried’ in it. Hence, the gene is ‘off’
In order to turn the gene ‘on,’ we can loosen the loops around the gene a bit so that it is more accessible for RNA polymerase, allowing the gene to be transcribed and expressed
Why is DNA replication so important?
important for how you age
big factor in cancer
important for how children are made
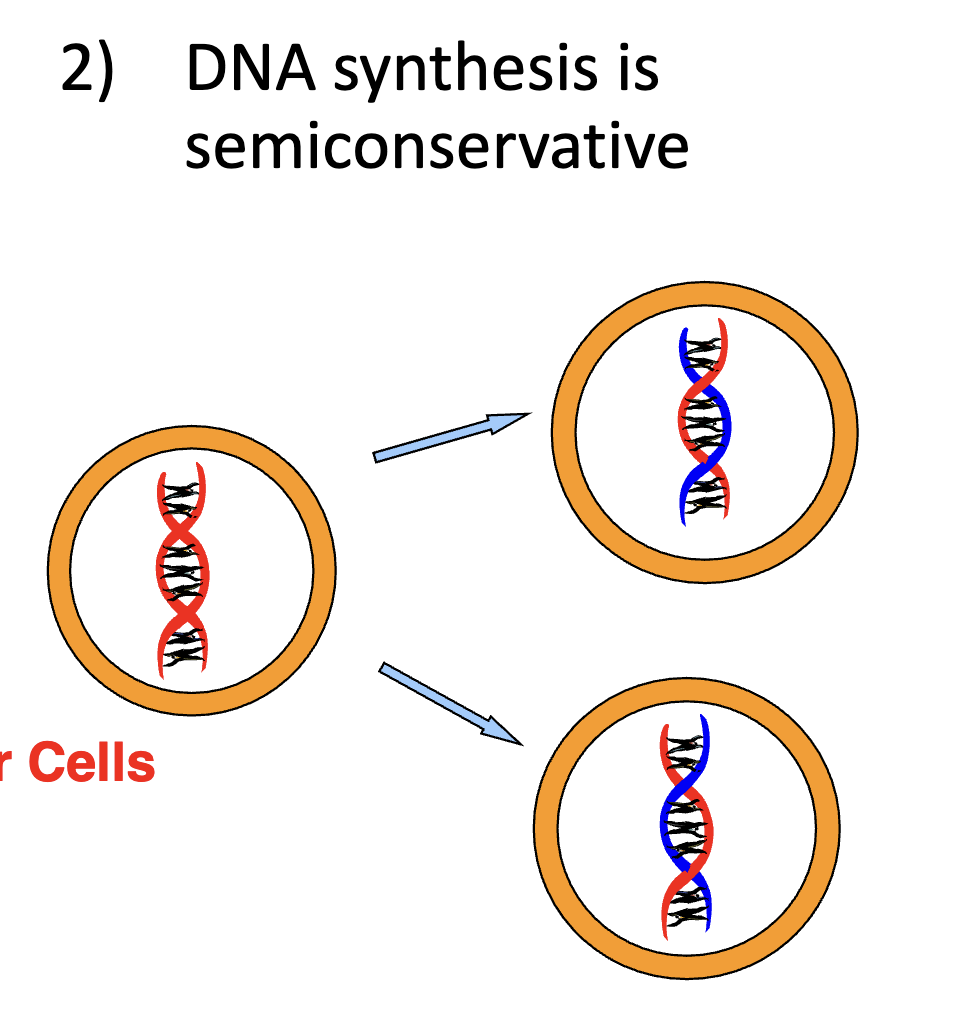
Is DNA conservative or semi-conservative?
Semi-conservative.
This means that one parent strand gets used in one daughter cell and the other parent strand is used in the other daughter cell. See image.
What is DNA synthesised from?
Deoxyribonucleoside Triphosphates (dNTPs).
What is RNA synthesised from?
Ribonucleoside Triphosphates (NTPs).
What links nucleotides?
Phosphodiester bonds.
What does DNA polymerase do?
It catalyses complementary base pairing.
What are the 3 main rules of DNA replication?
DNA is anti-parallel
New DNA is synthesised in the 5’ to 3’ direction (meaning you add onto a 3’ end)
The template strand is read in the 3’ to 5’ direction (so from the template strand perspective, the new strand is being made in the 3’ to 5’ direction)
Explain the model of DNA replication found in eukaryotes and bacteria.
Origin of Replication:
Bidirectional growth starts from one origin of replication. This means that from that point, the DNA double helix gets unwound, two replication forks are made (one side going left, the other side going right)
Right Side of Replication Fork:
On the right side of the replication fork, there are 2 exposed DNA strands. On those exposed DNA strands, newly synthesised DNA strands are made on each
These newly synthesised DNA strands are made in the 5’ to 3’ direction (so from the template strand perspective, the new strand is being made in the 3’ to 5’ direction)
On the leading strand, DNA is made in the direction of the separation. On the lagging strand, Okazaki fragments are made as new strands are made in the opposite direction of the separation
Left Side of Replication Fork:
Looking onto the left side of the DNA replication fork, the leading and lagging strand are now on different sides. This is because the 5’ and 3’ end on the template strands are on opposite sides.
In the image, on the left side of the replication fork, the Okazaki fragments are now on template strand A instead of on the right side of the replication fork where they were on template strand B.
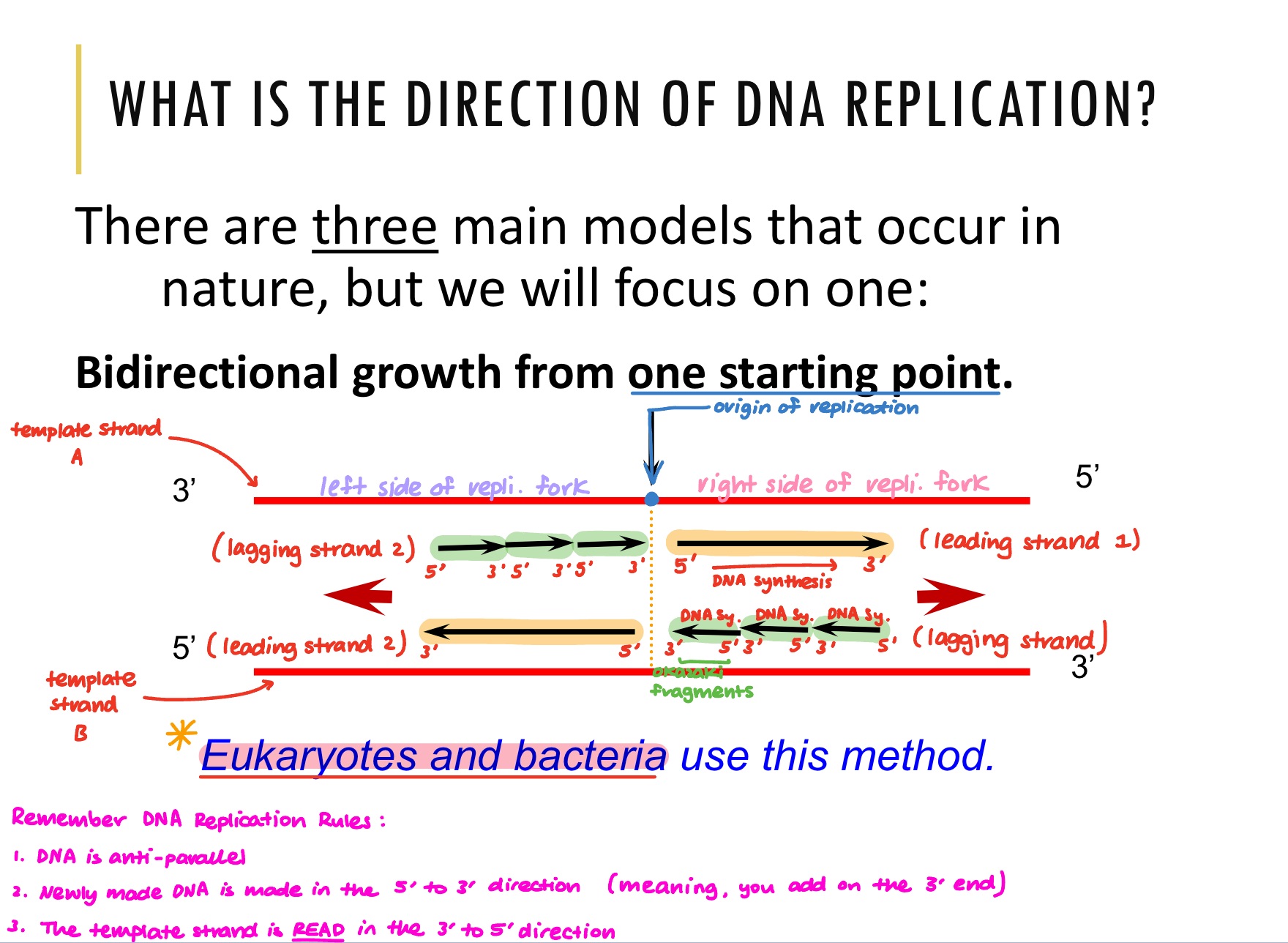
What does bidirectional DNA replication mean?
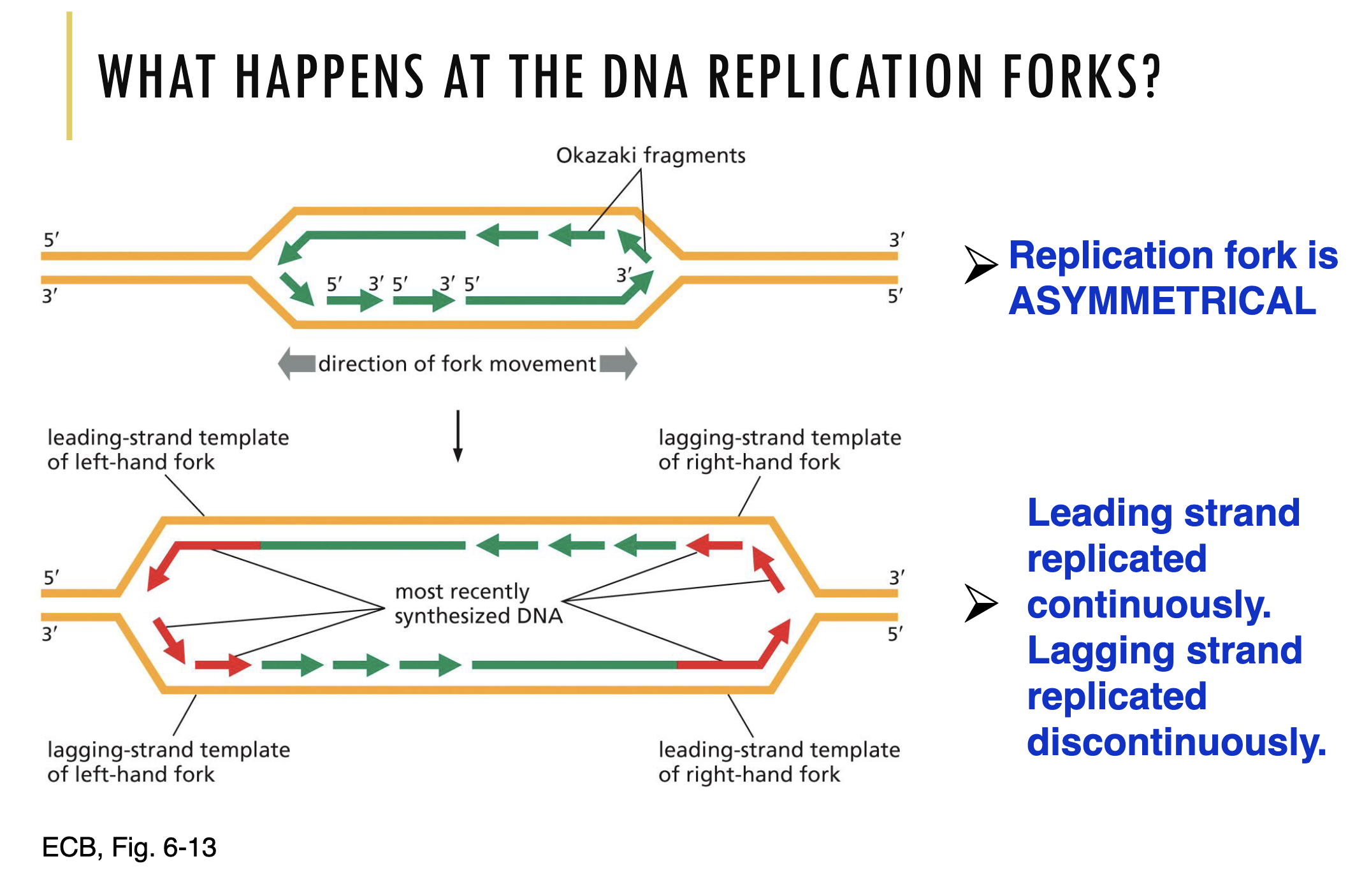
This means that at the origin of replication, two replication forks are made that are in opposite directions. Hence, DNA replication is taking place in 2 directions from one point of initiation. Refer to the video to visualise this.
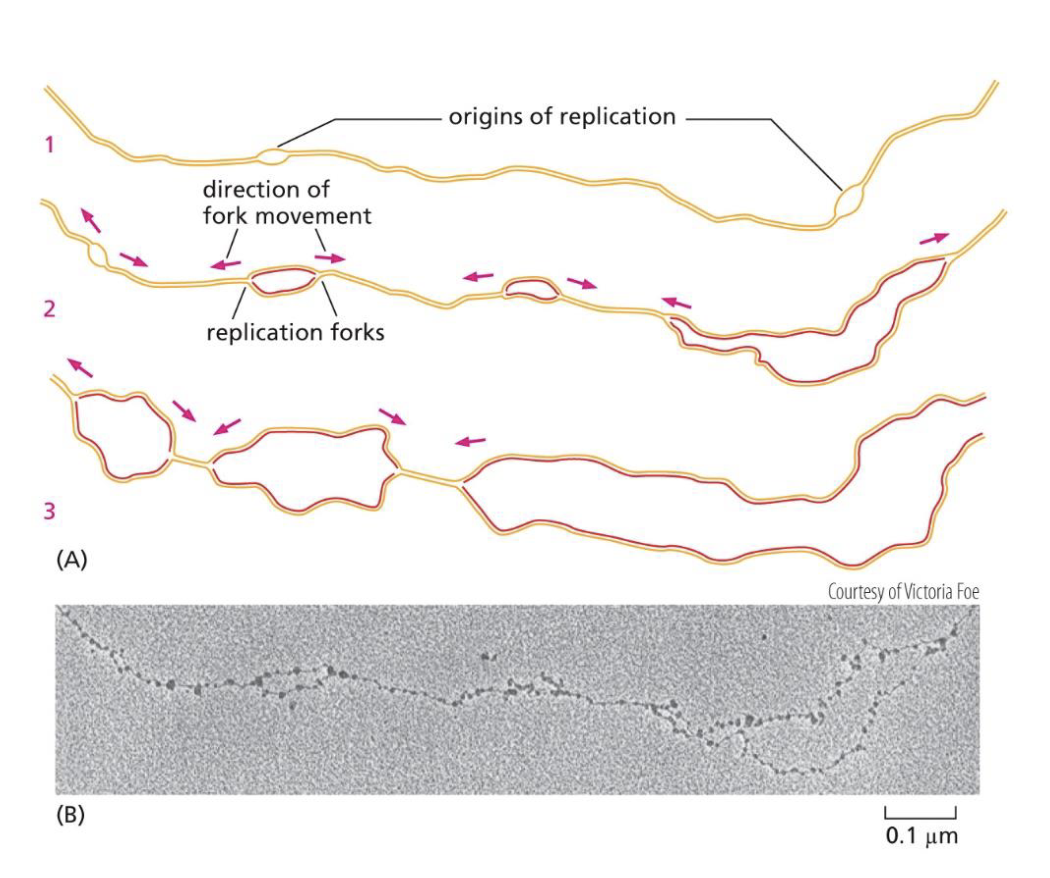
On a standard eukaryotic chromosome, are there multiple origins of replication? If so, what does that mean for bidirectional DNA replication?
Yes, there are multiple origins of replication on a standard eukaryotic chromosome.
That just means that at each point, bidirectional DNA replication occurs.
Where does DNA replication start?
DNA replication always starts from the location of DNA.
Hence, there are specific DNA sequences at replication origins that dictate where DNA replication should start.
What are the characteristics of the specific DNA sequences at replication origins?
The sequences are A-T rich as they are easy to open
The sequences are recognised by initiator proteins that bind to DNA
How many origins of replication do different types of organisms have?
Prokaryotes
single origin of replication
Eukaryotes
multiple origins of replication