Biol 3010 Exam II Review Flashcards
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
In a classic experiment reminiscent of Beadle and Tatum's seminal work, researchers studied a series of Neurospora crassa mutants to understand the genetic basis of amino acid synthesis. Each mutant was unable to synthesize the amino acid leucine due to a single gene defect. To determine which gene was affected in each mutant, the researchers conducted rescue experiments by adding intermediate compounds of the leucine biosynthesis pathway to the growth medium.
The following observations were made:
· Mutant A grew when the medium was supplemented with compound X or any subsequent intermediate in the pathway.
· Mutant B grew only when the medium was supplemented with leucine itself.
· Mutant C grew when the medium was supplemented with compound Y, compound Z, or leucine.
· Mutant D grew when the medium was supplemented with compound Z or leucine.
Assuming that the leucine biosynthesis pathway is linear and follows this sequence: Precursor → Compound W → Compound X → Compound Y → Compound Z → Leucine.
Which mutant has a defect in the gene encoding the enzyme responsible for the conversion of compound W to compound X?
Mutant A
Gene bodies sometimes overlap because:
exons and introns can be read from different strands of DNA
Which of the following promotes more efficient translation in the closed-loop mRNA structure?
Formation of a stable complex by mRNA bound PABP, eukaryotic translation initiation factors (eIF4A/eIF4E/eIF4G) and the 5’ cap
You sequence a sub-section of a larger gene. You identify the template strand sequence to be:
5’ - TAC CGG AAT AGC ACT -3’
What is the corresponding mRNA sequence of the template DNA?
5’-AGU GCU AUU CCG GUA-3’
You sequence a sub-section of a larger gene. You identify the template strand sequence to be:
5’ - TAC CGG AAT AGC ACT -3’
What is the corresponding protein sequence encoded by this DNA template strand? Assume the 1st position of the mRNA sequence is the first nucleotide of a codon in the protein?
N- Ser Ala Ile Pro Val -C
Which of the following are mechanisms known to regulate the function of transcription factors?
all of the above
In a patient with a rare metabolic disorder, researchers identified a missense mutation in the gene encoding the enzyme alanyl-tRNA synthetase (AlaRS). This mutation leads to a single amino acid change in the enzyme's active site, reducing its efficiency in charging tRNA molecules with alanine. As a result, some tRNAs that are normally charged with alanine are mischarged with glycine instead. Considering the genetic code and the structure of tRNA, this scenario primarily affects the translation of which codon(s) into alanine, and how might the missense mutation in the AlaRS gene influence protein synthesis in the patient's cells?
The mutation affects the translation of GCU, GCC, GCA, and GCG codons, which should specify alanine, potentially leading to the incorporation of glycine at positions where alanine is expected, altering protein structure and function.
How do histone acetyl transferase (HAT) enzymes promote gene expression?
by adding acetyl groups to specific amino acids in histone tails, reducing the affinity of nucleosomes for nearby DNA
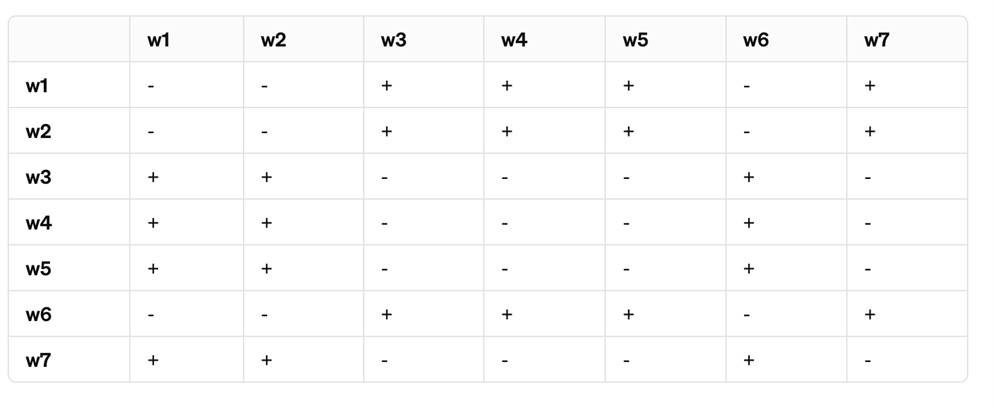
A team of geneticists is studying a set of seven Drosophila melanogaster mutants, all exhibiting a similar white eye phenotype. The mutants are labeled w1 through w8. To determine the genetic basis of the white eye phenotype, the researchers perform a series of complementation tests by intercrossing the mutants in all possible pairings. The following table summarizes the results, where “+” indicates the F1 progeny exhibit the wild-type red eye phenotype and “-” indicates the F1 progeny maintain the white eye phenotype.
From these results, identify the complementation groups among these mutants. How many complementation groups are there, and which mutants belong to each group?
There are 2 complementation groups "w1, w2, w6" belong to one group, "w3, w4, w5, and w7" belong to a second.
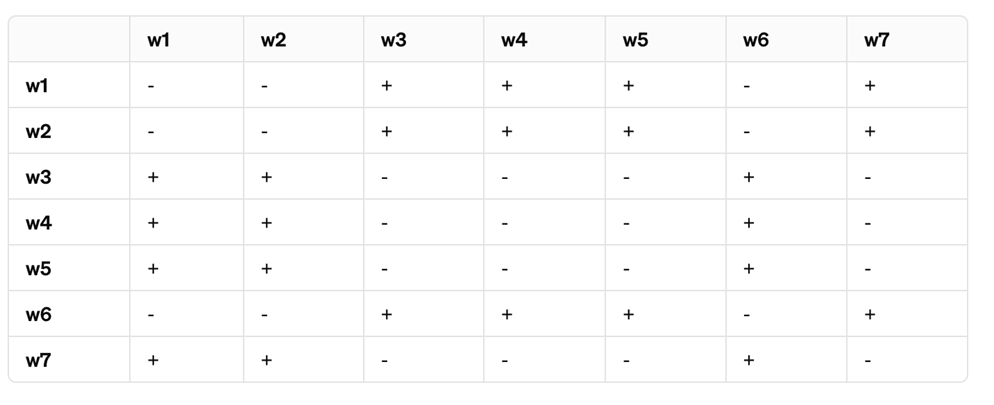
A team of geneticists is studying a set of seven Drosophila melanogaster mutants, all exhibiting a similar white eye phenotype. The mutants are labeled w1 through w8. To determine the genetic basis of the white eye phenotype, the researchers perform a series of complementation tests by intercrossing the mutants in all possible pairings. The following table summarizes the results, where “+” indicates the F1 progeny exhibit the wild-type red eye phenotype and “-” indicates the F1 progeny maintain the white eye phenotype.
Based on the above analysis, and given the fact that your screen was carried to saturation, how may genes control eye color in Drosophila?
(Saturation in genetic screening is the point where all recoverable genes affecting a phenotype have been mutated.)
2
pre-mRNA (aka the primary transcript) is post-transcriptionally processed to mature mRNA. All are important post-transcriptional modifications of pre-mRNA except:
removal of the 5'UTR so that the first codon is AUG
An Lys-tRNALys can recognize both AAA and AAG codons. Which anticodon sequence for this tRNA shows the Wobble position base in bold?
5’CUU3’
Where does alternative RNA splicing take place in eukaryotic cells and how is alternative splicing regulated?
Alternative splicing occurs in the nucleus, where SR proteins and hnRNPs interact with cis-acting enhancers and suppressors to regulate the inclusion or exclusion of exons
Eukaryotic genes have cis-regulatory regions sometimes referred to as enhancers. Why are enhancers important in gene regulation?
enhancers allow for gene expression to be controlled spatially and temporally
mRNAs with upstream open reading frames (ORFs) in their 5'UTRs typically have:
dissociation of 40S and 60S subunits before the main ORF initiator codon, thereby reducing translation of the main ORF.
After RNA pol II “escapes” from the Preinitiation Complex it:
synthesizes a short stretch of RNA complementary to the template strand of DNA before continuing through the gene body with the assistance of specific elongation factors
Pioneer transcription factors differ from most transcription factors in being able to
all of the above
What is a potential molecular consequence of a single nucleotide change within the coding sequence or regulatory element of a gene?
all of the above
Ribosomopathies are diseases caused by defects in ribosome biogenesis or function. Some ribosomopathies primarily affect specific cell types despite ribosomes being essential in all cells. Which of the following best explains why certain ribosomopathies selectively impact rapidly dividing cells, such as erythroid progenitors in Diamond-Blackfan anemia?
Rapidly dividing cells have a higher demand for ribosome production, so defects in ribosome biogenesis disproportionately affect their ability to synthesize proteins.
In the Yanofsky experiment, how could different mutations in the DNA sequence result in different amino acids at the exact same location in TrpA after translation?
I. the mutations affect the start codon, methionine
II. the mutations were at the same nucleotide position
III. the mutations were at different nucleotide positions but within the same codon triplet
IV. the mutations were at different splice sites
II and III
You and your colleague are interested in identifying genes that control organism growth. In a large scale EMS mutagenesis screen, you identified 40 mutants. These 40 mutants each contain a mutation that when homozygous gives rise to animals that are smaller in size compared to control groups. Which of the following statements is correct regarding the nature of your screen and the mutants you have identified?
I. I expect that all mutants will have mutations in the coding region of genes important for growth control.
II. I expect that the lesions in general will be G:C to A:T transitions.
III. I expect that all 40 mutations will be in unique genes.
IV. If all 40 mutations are indeed in unique genes, I can safely conclude that I have identified all genes important for growth control.
II
The fictitious trx gene’s protein coding sequence is 297 base pairs (bp) long. Which of the following is the most likely to affect the phenotype of the organism that expresses this gene?
a nonsense mutation in the 39th nucleotide of the DNA sequence
The polyA tail of mRNA:
I. signals the site of transcriptional termination by marking the GU-rich endonuclease cut site
II. can code for poly-lysine (AAA) protein tails (i.e. in histones)
III. protects the transcript from degradation
IV. enhances translation efficiency
III and IV
The classical "Central Dogma" of molecular biology states that:
none of the above
Which of the following is true about cis-regulatory elements:
I. They are difficult to predict and can be found 5’ to the basal promoter as well as in introns
II. They are DNA sequences that bind protein
III. They are often translated at the same time as mRNA
IV. Eukaryotic genes typically are controlled by only one cis-regulatory sequence
I and II
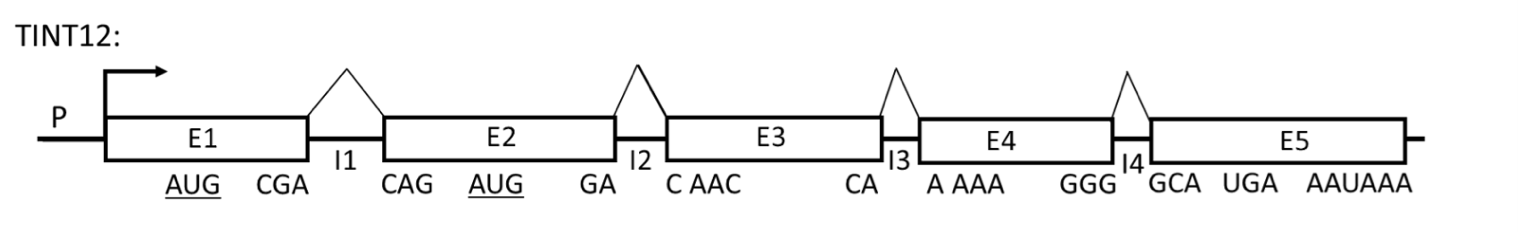
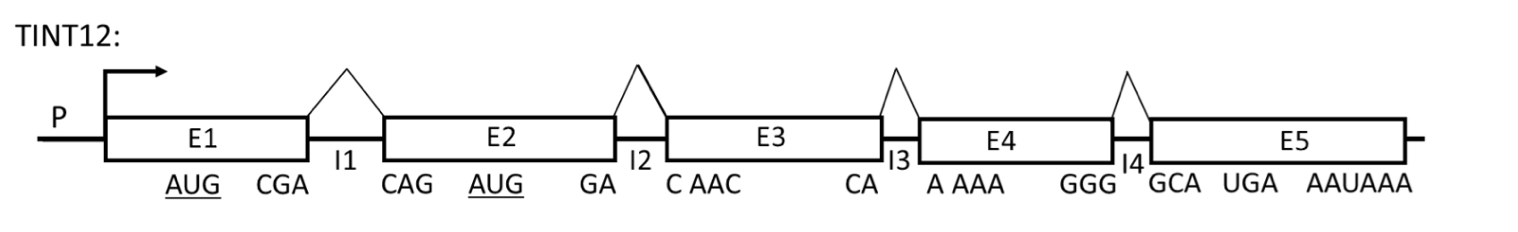
Refer to the figure below of a hypothetical gene (TINT12) to answer the following 4 questions.
Promoter is P; E1, E2, E3, E4 and E5 are exons; I1, I2, I3 and I4 are introns (the components are not drawn to scale). The positions of the codons at each exon/intron junction in the reading frame established by the AUG codon in E1 (AUG-E1) are indicated below each exon. A codon encoding Met in the second exon (AUG-E2) is in the same reading frame as AUG-E1. The stop codon and the poly(A) signal are indicated.
In normal cells, E1E2E3E4E5 mRNA is not very abundant even though the promoter is very efficient. E1E2E3E4E5 mRNA has a half life of only 30 minutes. 90% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE1. 10% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE2.
As a reminder, the optimal Kozak sequence is (gcc)RccAUGG, where R is a purine (A or G) at the -3 position relative to the AUG start codon, and G is highly favored at the +4 position while the optimal 5’-end of an intron sequence is 5’GU (donor site) and the 3’-end is 5’AG (acceptor site).
In a comparative study between the normal cell line and one expressing a mutant version of the TINT12 gene (mutA), analysis revealed that both cell lines produce the mRNA transcript E1E2E3E4E5 at comparable levels. However, a significant shift in translation initiation was observed in the mutant cell line: 90% of the synthesized protein began with an N-terminal methionine encoded by the AUG codon in exon E2 (AUGE2), whereas only 10% utilized the AUG codon in exon E1 (AUGE1) for translation initiation. Given the sequences surrounding these AUG codons, known as Kozak sequences, which of the following best explains the altered preference for the start codon in the mutant TINT12 gene (mutA)?
Original Kozak sequence in E1: ACCAUGGG; E2: ACCAUGGG
Mutant Kozak sequence in E1: CCCAUGGG; E2: ACCAUGGG
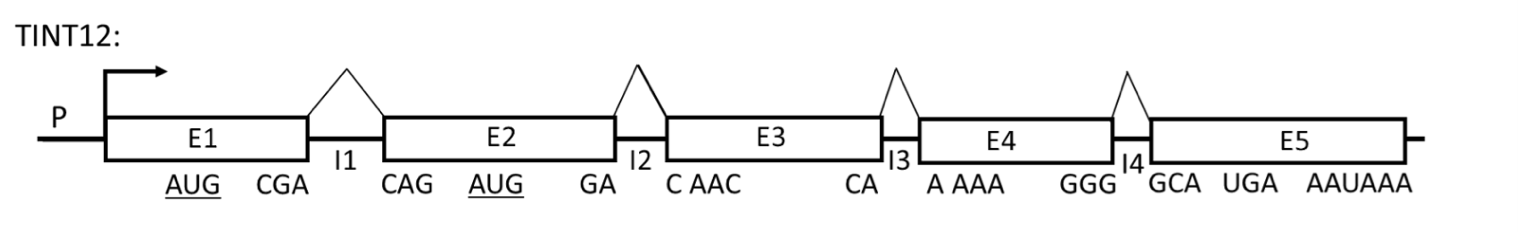
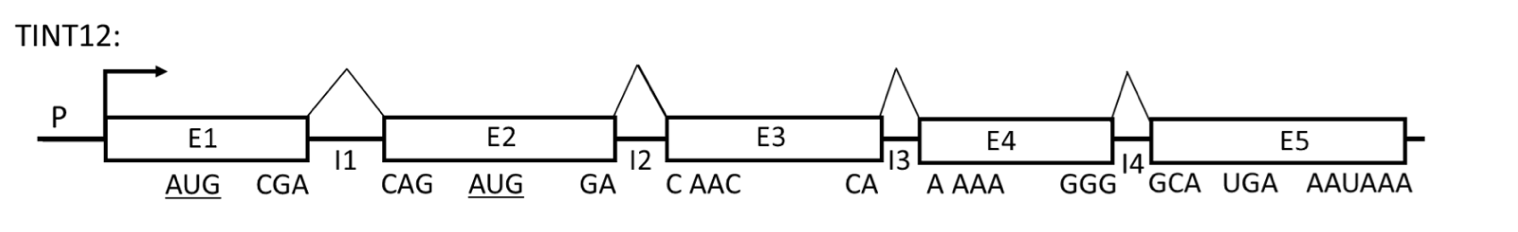
Refer to the figure below of a hypothetical gene (TINT12) to answer the following 4 questions.
Promoter is P; E1, E2, E3, E4 and E5 are exons; I1, I2, I3 and I4 are introns (the components are not drawn to scale). The positions of the codons at each exon/intron junction in the reading frame established by the AUG codon in E1 (AUG-E1) are indicated below each exon. A codon encoding Met in the second exon (AUG-E2) is in the same reading frame as AUG-E1. The stop codon and the poly(A) signal are indicated.
In normal cells, E1E2E3E4E5 mRNA is not very abundant even though the promoter is very efficient. E1E2E3E4E5 mRNA has a half life of only 30 minutes. 90% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE1. 10% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE2.
As a reminder, the optimal Kozak sequence is (gcc)RccAUGG, where R is a purine (A or G) at the -3 position relative to the AUG start codon, and G is highly favored at the +4 position while the optimal 5’-end of an intron sequence is 5’GU (donor site) and the 3’-end is 5’AG (acceptor site).
Research into the TINT12 gene has unveiled that a mutant variant, referred to as mut$, possesses a 40 base pair deletion within its third intron (I3). Subsequently, it has been observed that the E1E2E3E4E5 mRNA transcribed from mut$ not only exhibits a marked increase in abundance but also demonstrates an extended half-life, persisting for 24 hours. Which of the following explanations is the most plausible for the observed changes in mRNA stability and abundance in the mut$ cells?
The intact wild-type intron I3 contains a sequence that is processed into a functional miRNA; this miRNA has complete complementarity to a segment in one of the exons of TINT12.
Refer to the figure below of a hypothetical gene (TINT12) to answer the following 4 questions.
Promoter is P; E1, E2, E3, E4 and E5 are exons; I1, I2, I3 and I4 are introns (the components are not drawn to scale). The positions of the codons at each exon/intron junction in the reading frame established by the AUG codon in E1 (AUG-E1) are indicated below each exon. A codon encoding Met in the second exon (AUG-E2) is in the same reading frame as AUG-E1. The stop codon and the poly(A) signal are indicated.
In normal cells, E1E2E3E4E5 mRNA is not very abundant even though the promoter is very efficient. E1E2E3E4E5 mRNA has a half life of only 30 minutes. 90% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE1. 10% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE2.
As a reminder, the optimal Kozak sequence is (gcc)RccAUGG, where R is a purine (A or G) at the -3 position relative to the AUG start codon, and G is highly favored at the +4 position while the optimal 5’-end of an intron sequence is 5’GU (donor site) and the 3’-end is 5’AG (acceptor site).
In the TINT12 gene, a variant referred to as mutV exhibits a silent mutation within exon 3, altering a GCA codon to a GCG codon. Surprisingly, this single nucleotide change leads to a drastic 90% decrease in the production of the functional TINT12 protein. What is the most plausible explanation for the significant reduction in protein yield despite the mutation being silent?
The silent mutation in E3 of TINT12-mutV creates a cryptic splicing site, leading to aberrant splicing and the production of a mis-spliced mRNA transcript.
Refer to the figure below of a hypothetical gene (TINT12) to answer the following 4 questions.
Promoter is P; E1, E2, E3, E4 and E5 are exons; I1, I2, I3 and I4 are introns (the components are not drawn to scale). The positions of the codons at each exon/intron junction in the reading frame established by the AUG codon in E1 (AUG-E1) are indicated below each exon. A codon encoding Met in the second exon (AUG-E2) is in the same reading frame as AUG-E1. The stop codon and the poly(A) signal are indicated.
In normal cells, E1E2E3E4E5 mRNA is not very abundant even though the promoter is very efficient. E1E2E3E4E5 mRNA has a half life of only 30 minutes. 90% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE1. 10% of the protein produced from E1E2E3E4E5 mRNA has an N-terminal Met encoded by AUGE2.
As a reminder, the optimal Kozak sequence is (gcc)RccAUGG, where R is a purine (A or G) at the -3 position relative to the AUG start codon, and G is highly favored at the +4 position while the optimal 5’-end of an intron sequence is 5’GU (donor site) and the 3’-end is 5’AG (acceptor site).
A mutant version of TINT12 (mut2) has a complete deletion of E2. From the list below please select the exon(s) you would have to “skip” to generate an internally truncated, but potentially in-frame functional protein?
E3+4
The following partial mRNA sequence encodes the first seven amino acids of a eukaryotic protein:
5'-AUG GCU ACU UGG AAA GGC GGA-3'
Three mutations are introduced resulting in the addition of nucleotides (A, G, and U) at different locations:
An A is inserted after the first codon, which is also the start (AUG → AUGA).
A G is inserted between the second and third nucleotides of the third codon (ACU → ACGU).
A U is inserted between the first and second nucleotides of the fifth codon (AAA → AUAA).
Which of the following best describes the consequence of these mutations?
The insertions introduce a premature stop codon leading to a truncated protein