BIOL 3001 Final
1/129
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
130 Terms
DNA
double helix molecule made up of nucleotides, which are the building blocks of genetic information. The sequence of these nucleotides encodes the instructions for building and maintaining an organism.
Genome
encompasses all the DNA within an organism, including all its genes and the non-coding DNA that supports them. It's like the complete instruction manual for an organism.
Human Genome Prokect
a massive, international scientific endeavor to map and sequence the entire human genome. It aimed to identify all the genes within human DNA and make the resulting data publicly available for research purposes.
Bioinformatics
an interdisciplinary field that combines biology, computer science, and information technology to analyze and interpret biological data
non-coding DNA
the portion of an organism's DNA that does not code for proteins. While historically considered inactive or insignificant, it's now understood to play crucial roles in regulating gene expression and other cellular processes.
Sequence inspection
Can be used to locate genes because genes are not simply a random series of nucleotides but instead have distinctive features
Not a foolproof way of locating genes, but still a powerful tool and is usually the first method that is applied to analysis of a new genome sequence
ORF scanning
Effective way of locating most, but not all of the genes in the DNA sequence
Analysis of bacterial sequences is simplified = don't have non-coding introns
Intergenic
stretches of DNA sequences located between genes. These regions can be involved in regulating gene expression and may contain functional elements or "junk DNA". They are also referred to as spacer DNA
Codon bias
not all codons are used equally frequently in the genes of a particular organism
Exon-intron boundaries
Semi-distinct sequence features
Upstream regulatory sequences
DNA sequences located before (or upstream of) a gene's coding region that control the gene's expression
CpG islands
a region of DNA with a high frequency of CpG dinucleotides (cytosine followed by guanine, linked by a phosphate group). These islands are typically found near the promoter regions of genes and play a crucial role in regulating gene expression
GenBank
A database of previously sequenced and identified genes available through the NCBI
designed to provide and encourage access within the scientific community to the most up-to-date and comprehensive DNA sequence information
cDNA
Complementary DNA
DNA copy of an mRNA molecule
Used in GenBank to represent the coding sequences of genes, particularly when studying gene expression or when cloning eukaryotic genes into prokaryotic systems
Lacks introns and only includes expressed regions of a gene
Reverse transcriptase
an enzyme that synthesizes DNA from an RNA template, a process called reverse transcription
cDNA library
A limited gene library using complementary DNA. The library includes only the genes that were transcribed in the cells examined.
reflects the gene expression profile of a specific cell or tissue
Assembly
taking the large numbers of generated DNA sequences and finding areas of overlap between them
De Novo Sequencing
sequencing a novel genome where there is no reference sequence available for alignment.
Fragmenting
Original DNA is broken into smaller pieces, often randomly
Sequencing
Fragments are sequenced, generating a large number of short DNA sequences ("reads")
Alignment and merging
The reads are aligned and merged based on the overlapping sequences to create larger, contiguous sequences called contigs
Scaffolding and ordering
Contigs are ordered and oriented using additional information, and then potentially ordering the scaffolds into chromosomes
Annotation
The assembled sequence is annotated, identifying genes and other functional elements
Gibson assembly
a molecular cloning method used to join multiple DNA fragments together in a single, isothermal reaction
powerful technique known for its efficiency, speed, and ability to assemble large DNA fragments
Needs three enzymes: 5' exonuclease, a DNA polymerase, and a DNA ligase
Alignment
performed when the new DNA sequence generated is compared to existing DNA sequences to find any similarities or discrepancies between them and then arranged to show these features
Basic Local Alignment Search Tool (BLAST)
refers to a suite of programs used to generate alignments between a nucleotide or protein sequence, referred to as a "query" and nucleotide or protein sequences within a database, referred to as "subject" sequences
Alignment score
Numerical value reflecting the quality of the match
Percent identity
How many of the nucleotides are identical between the query and target sequences
E-value
A statistical measure of how likely it is to find a match of this quality purely by chance
Gene prediction
the process of identifying the regions of genomic DNA that encode genes
Evidence based prediction
Relies on previously gathered evidence regarding specific DNA sequences
TATAA box
thymine and adenine rich region of a gene located in most eukaryotic promoter regions
Located 10 bp upstream of the transcription initiation start site
5' UTR
Helps initiate transcription
3' UTR
Aids in transcription termination
Exons
Contain coding information that will be translated
Introns
Found between exons and are spliced or removed from the transcript before translation
Poly-A tail
Series of adenines are added to the 3' end of the mRNA
Start codon
AUG
Signals where protein synthesis should start
Stop codon
UAA, UAG, UGA
Signal protein synthesis to stop
Genome browser
software tool used to visualize and explore genomic data, including DNA sequences and annotations like genes, regulatory elements, and other features
Electrophoresis
technique commonly used in the laboratory to separate charged macromolecules, such as DNA or protein, according to size
may also be based on molecular weight, length and/or structure (shape)
Migration
Movement of charged molecules through the matrix in electrophoresis
DNA sequencing
the process of determining the order of nucleotides (adenine, guanine, cytosine, and thymine) within a DNA molecule
Chain termination sequencing
works by synthesizing new DNA strands complementary to a template, but the process is terminated when a dideoxynucleotide. (ddNTP) is incorporated instead of a normal deoxynucleotide (dNTP). This creates fragments of varying lengths, which are then separated by gel electrophoresis and analyzed to reveal the DNA sequence.
Sanger sequencing
a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication
DNA template
the DNA fragment that is being sequenced.
single-stranded DNA.
Thousands of copies of each fragment are required.
These copies can be created by cloning but most methods now use template DNA fragments produced using the Polymerase Chain Reaction.
This technique allows a researcher to target very specific DNA locations within the genome and amplify millions of near-perfect copies within a few minutes or hours.
DNA primer
a short strand of DNA that binds to the DNA template near the beginning of the fragment being sequenced
Deoxynucleotides
All four of the normal deoxynucleotides are included in the DNA synthesis reactions. The deoxynucleotide triphosphates are abbreviated dATP, dCTP, dGTP and dTTP, depending on which of the four different nitrogenous bases are attached (adenine, cytosine, guanine or thymine). When a nucleotide is added to a growing strand, two phosphates are released. The energy from this release drives the formation of the phosphodiester bond between the incoming nucleotide and the primer end.
DNA polymerase
required to catalyze formation the phosphodiester bonds. Polymerases used in sequencing are usually modified to operate efficiently in test tube environments and to have excellent proofreading capabilities (error prevention)
Reaction buffer
various chemicals and salts are added to highly purified water to mimic conditions in the cell where Replication normally occurs.
Di-deoxynucleotides
identical to the deoxynucleotides normally used during DNA Replication, with the exception that they lack the OH group on the 3' position of the ribose sugar.
During DNA sequencing, a di-deoxynucleotide can be added to a growing DNA strand but because the 3' OH is missing, the next phosphodiester bond cannot form (i.e. chain termination occurs).
Where should I begin my Sanger sequencing read?
Because the smallest fragments travel further in the gel than larger fragments, the DNA fragment at the very bottom of the gel is where you should begin. This fragment is a single "faulty" nucleotide attached to the primer you used in the sequencing reaction. IF the primer were 10 bases in length, the fragment at the bottom of the gel would be 11 bases long.
Which base am I looking at on the DNA fragment in sanger sequencing?
Remember, you ran FOUR different reactions, each containing a single and different di-deoxynucleotide. Those are the samples loaded in each lane. Read the nucleotide at the top of gel to know which one was added.
Chromatography
a laboratory technique used to separate the components of a mixture.
Chromatogram
a visual representation of the separation of components (DNA) in a mixture achieved through chromatography. It is essentially a graph where the signal intensity from a detector is plotted against time (or elution volume). Each peak on the chromatogram corresponds to a different component of the mixture, in this case, a different strand of DNA.
Genotyping
the process of determining the specific genetic makeup, or genotype, of an individual.
Microsatellite genotyping
a molecular biology technique used to identify and analyze variations within microsatellites, also known as short tandem repeats (STRs) or simple sequence repeats (SSRs), in DNA
Polymorphic
the state or quality of having or existing in multiple forms; the microsatellite region across individuals in a population will vary significantly
Linkage maps
a representation of the relative positions of genes or genetic markers on a chromosome, based on how frequently they are inherited together
Forward and reverse primer
Short, single stranded DNA sequences that bind to specific regions on a DNA template, flanking the target DNA sequence to be amplified. The forward primer binds to the antisense (or template) strand of DNA, while the reverse primer binds to the sense (or non-template) strand
Homozygotes appear as
a SINGLE band
Heterozygotes appear as
two bands
DNA profiling
the process where a specific DNA pattern, called a profile, is obtained from a person or sample of bodily tissue
Karyogram
shows the chromosomes of an organism in homologous pairs of decreasing length.
Genic DNA
~20% of the total genome
composed of the actual "coding regions" of DNA, those regions that provide the blueprint and information to synthesis proteins in the cells of the body
Also comprised of all the regulatory regions of the DNA that controls the synthesis of proteins. Regions of DNA like promoters, enhancers, repressors or polyadenylation signals are some examples of regulatory regions.
Extragenic DNA
~80% of the total genome
has non-coding regions of DNA. In the past, it has been classified as "Junk DNA" and thought to have no functionality
About half (~50%) of extragenic DNA is composed of what is termed "Repetitive DNA". A simple definition of this type of DNA are sequences that are present in multiple copies within a genome
Tandem repeats
sequences of DNA where a single unit of one or more base pairs is repeated multiple times in a tandem or head-to-tail fashion. These include all the "satellite regions" of the genome (minisatellites, microsatellites, alpha satellites, etc.).
Interspersed repeats
sequences of DNA that are repetitive in nature but are scattered throughout the genome rather than being clustered together like satellite regions. These include elements like interspersed repeats, long terminal repeats and transposons.
Polymorphic regions
some specific regions that are highly variable between individuals
Short Tandem Repeats (STRs)
used to create a DNA profile that is highly unique to each individual person.
Aka microsatellites
Regions of non-coding DNA that contain repeats of the same nucleotide sequence
Electropherograms
A graphical representation of STRs located on a person's chromosomes used to identify and tie suspects to a crime scene
Female (pedigree)
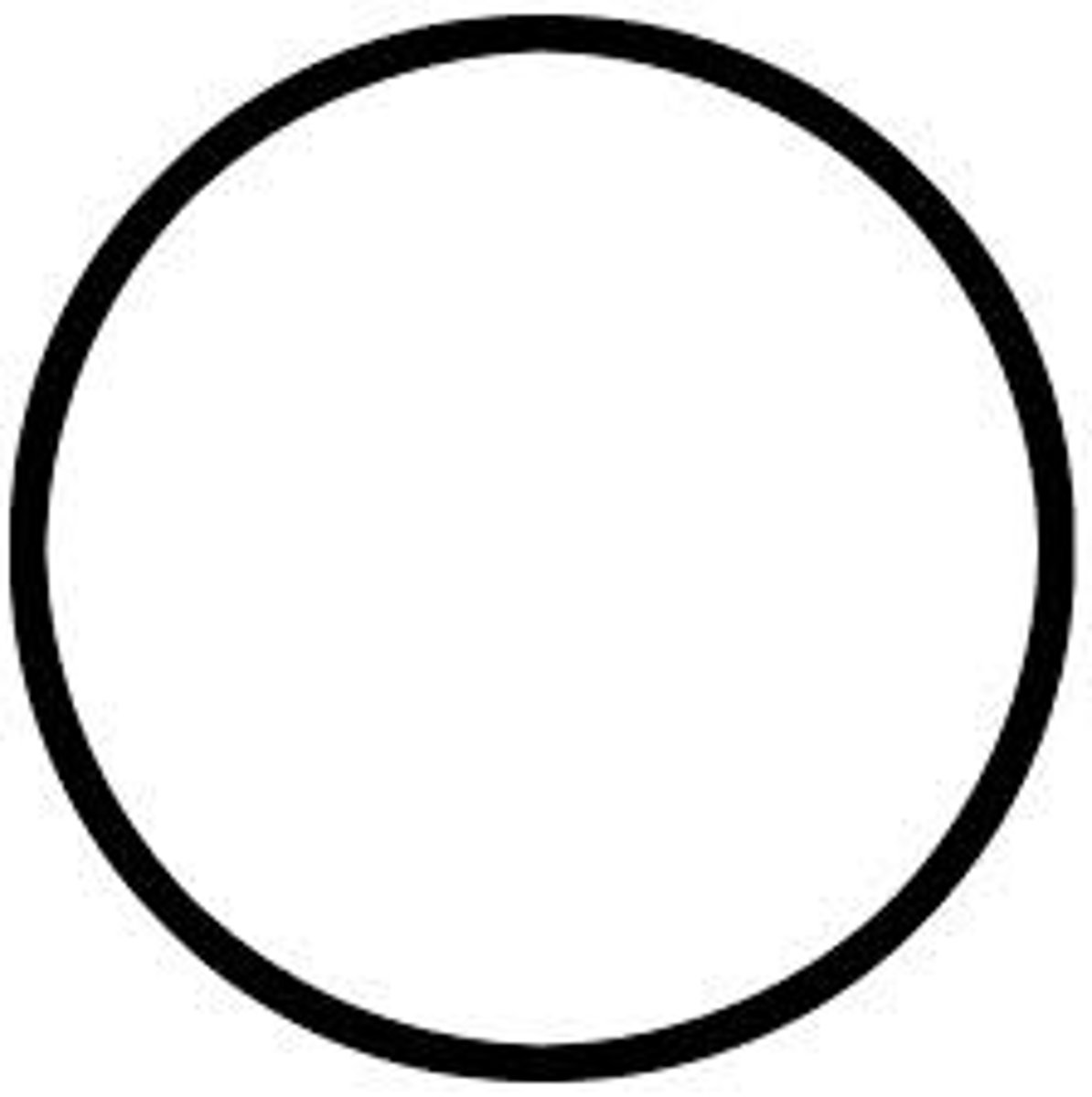
Male (pedigree)

Affected individuals

Carriers

Dead (pedigree)
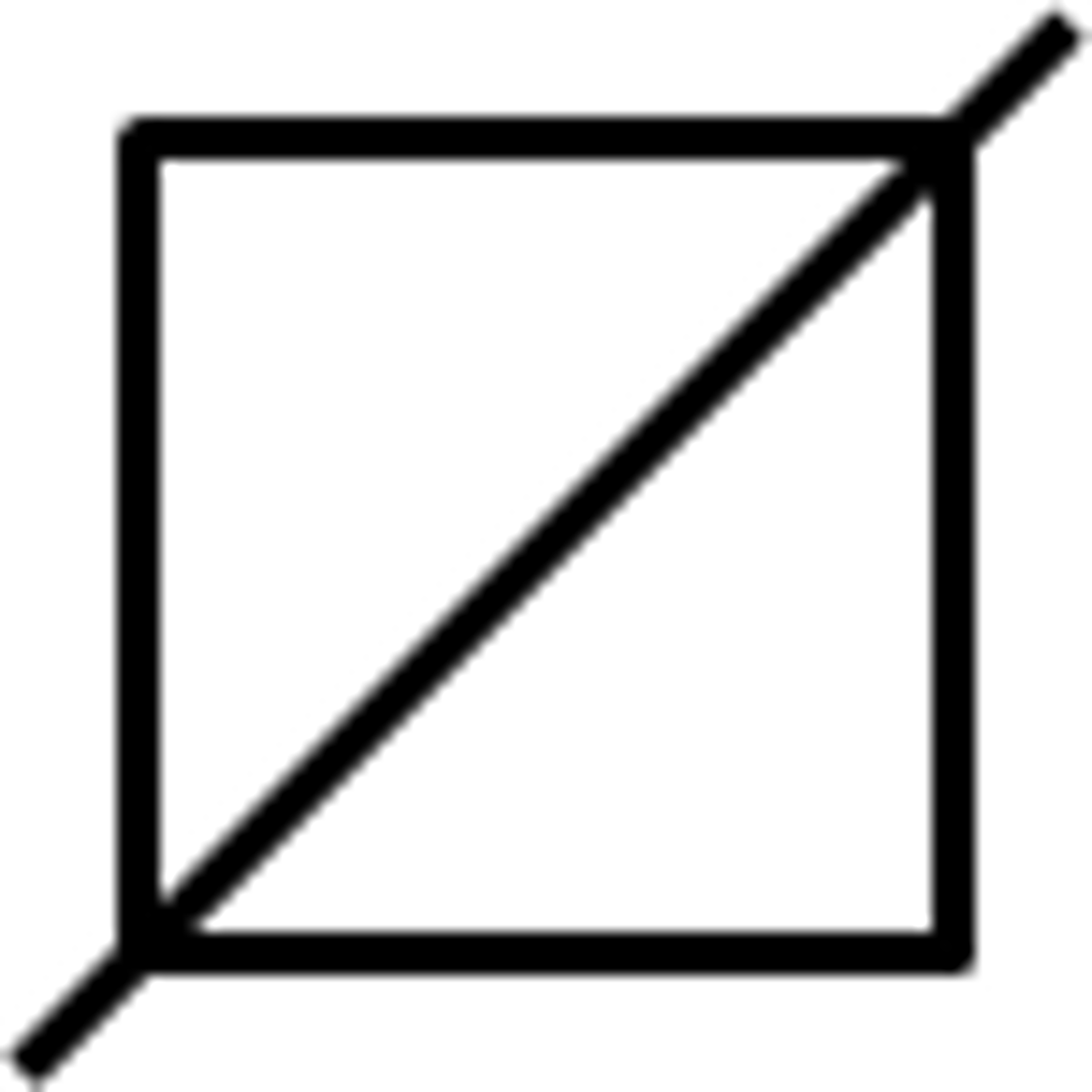
Gender not specified
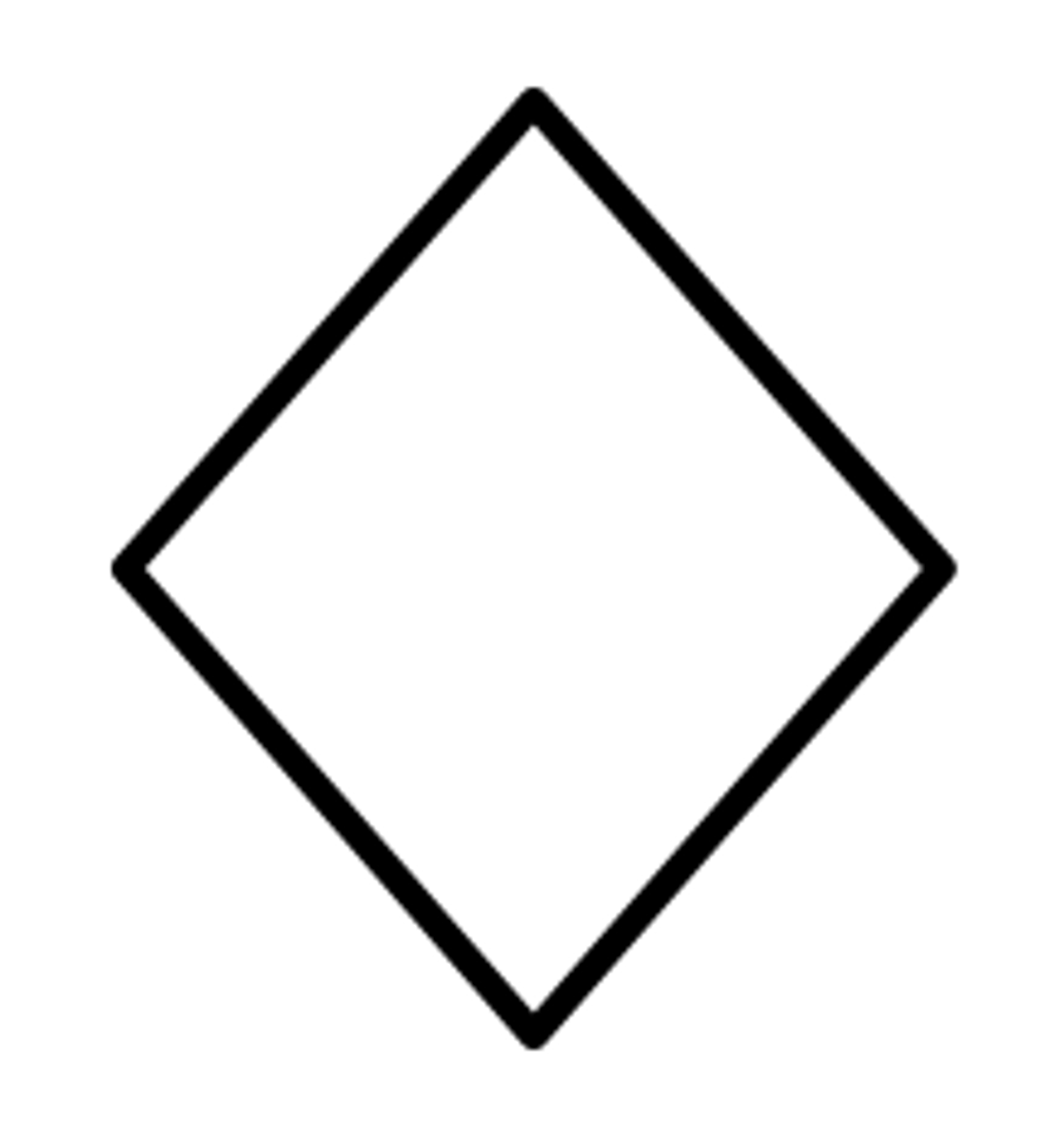
Spontaneous abortion (miscarriage)
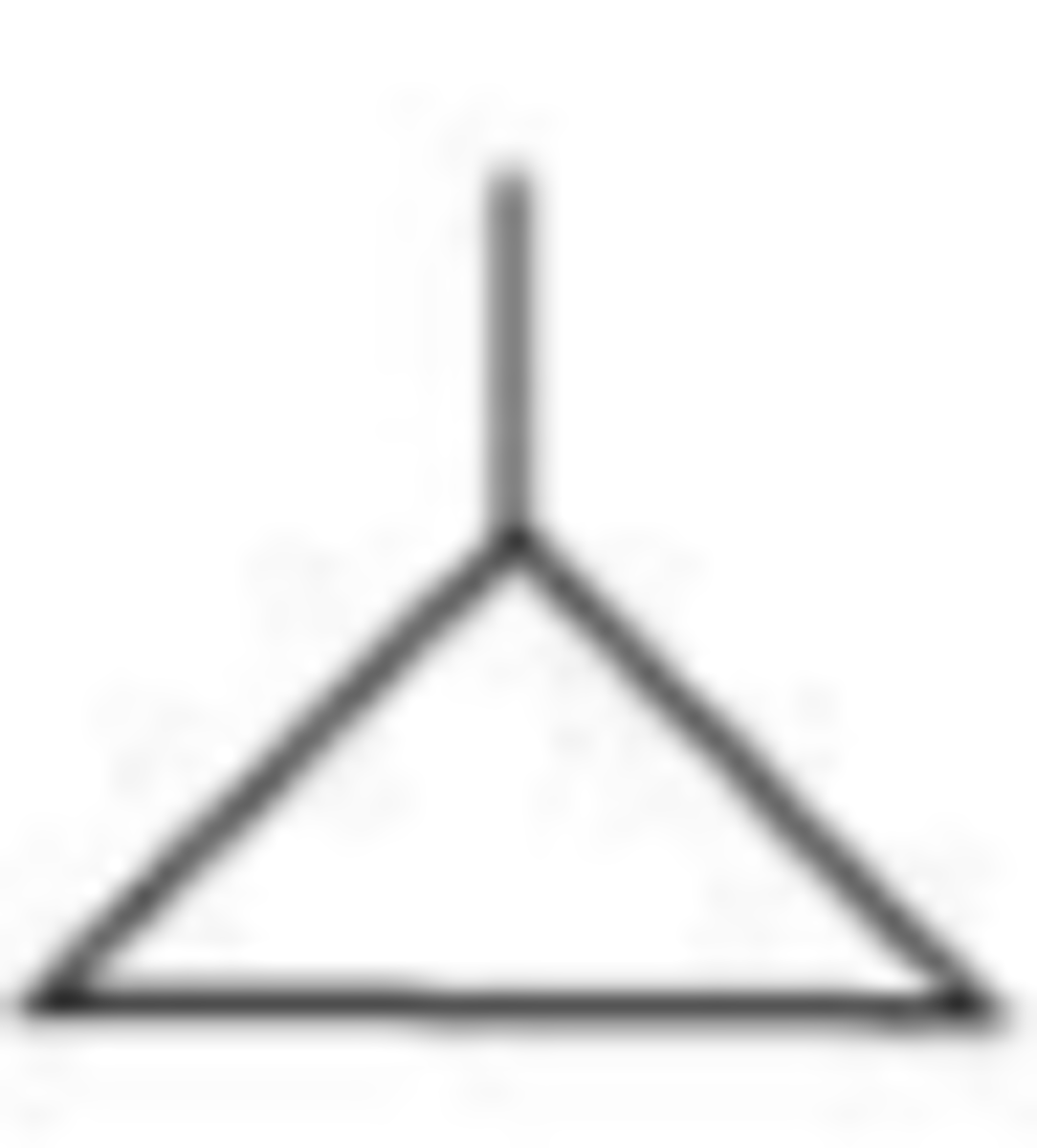
Still born
Terminated pregnancy
dizygotic twins (fraternal)
Monozygotic twins (identical)
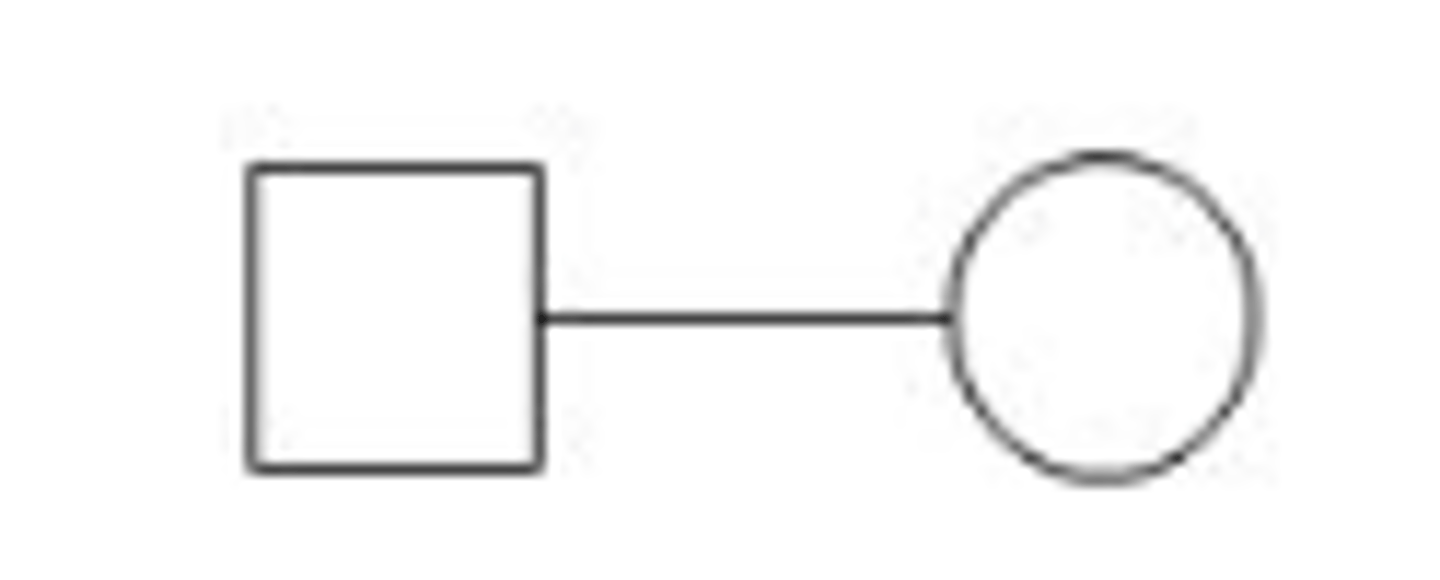
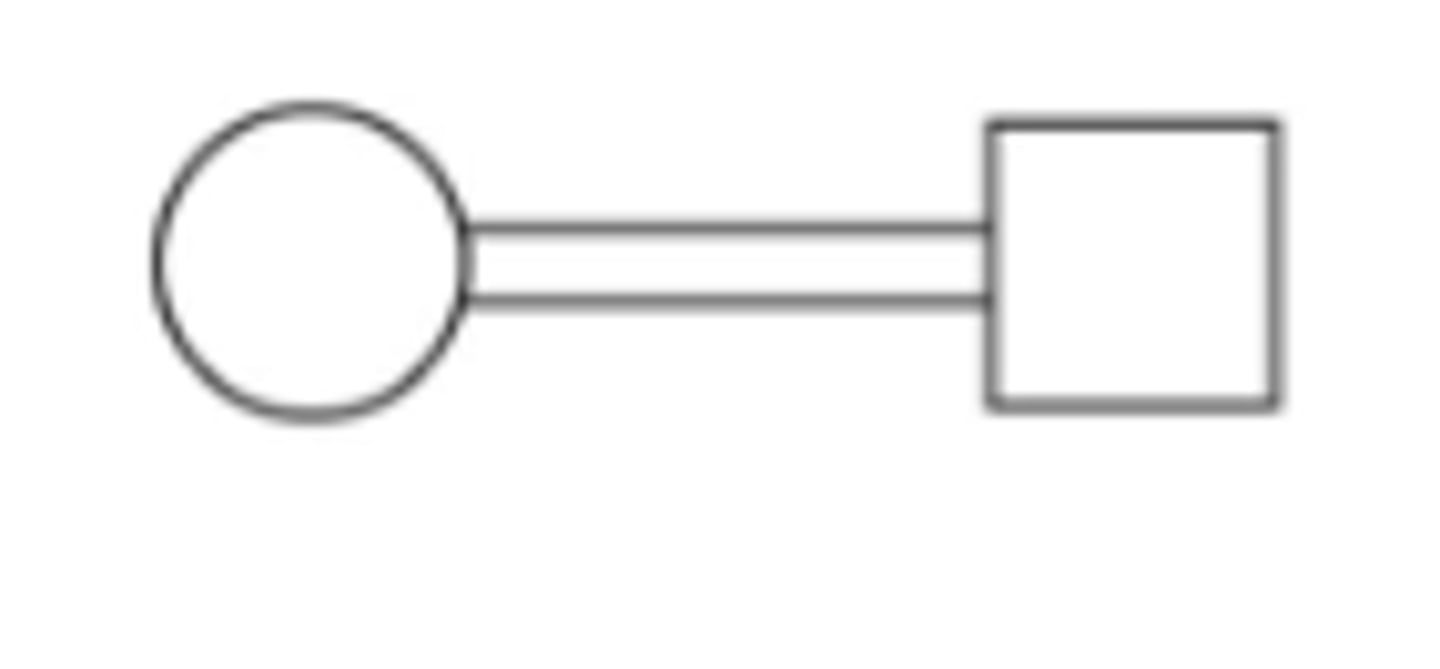
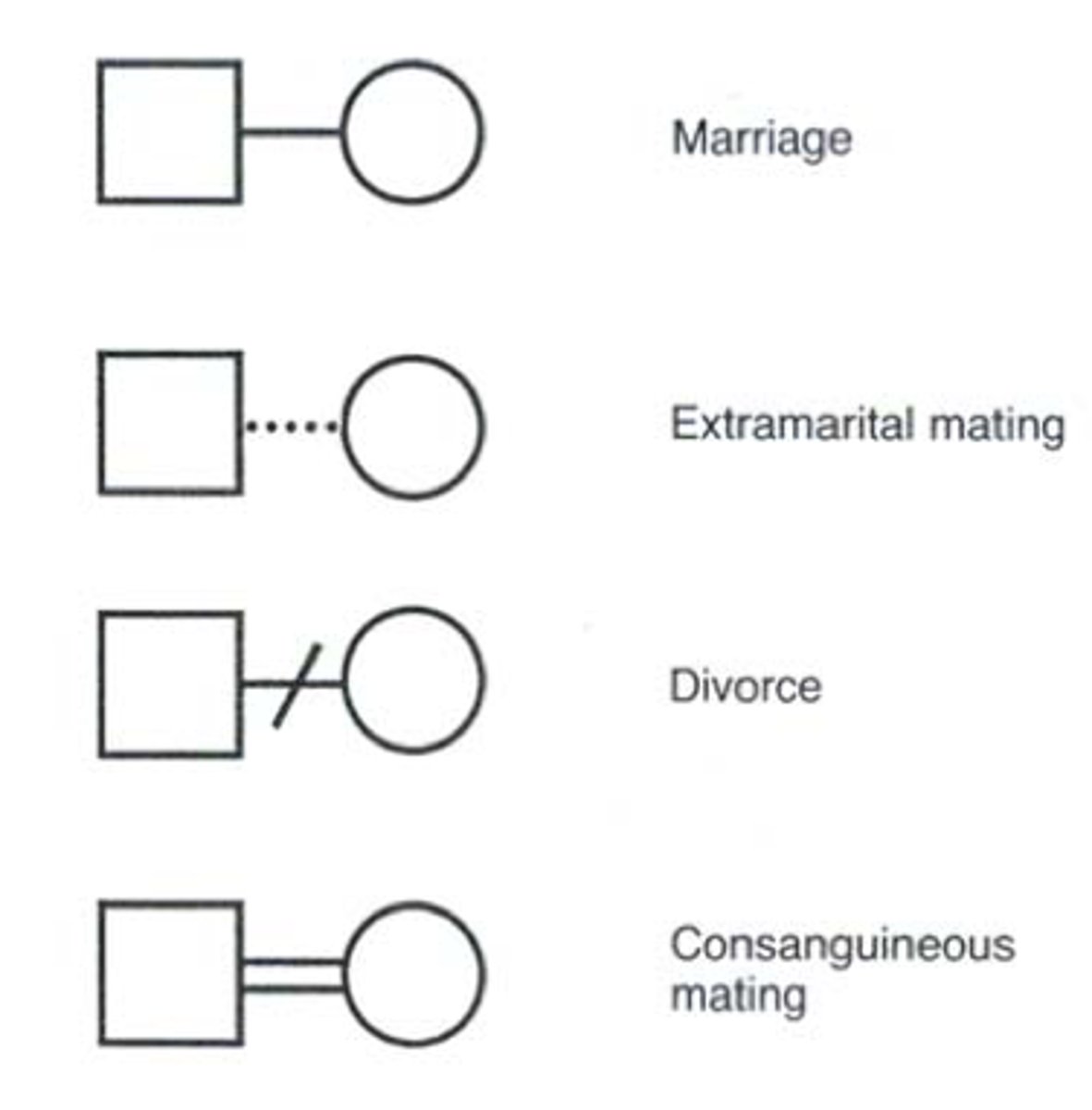
Mating
Consanguinous mating (inbreeding)
Extramarital relationship
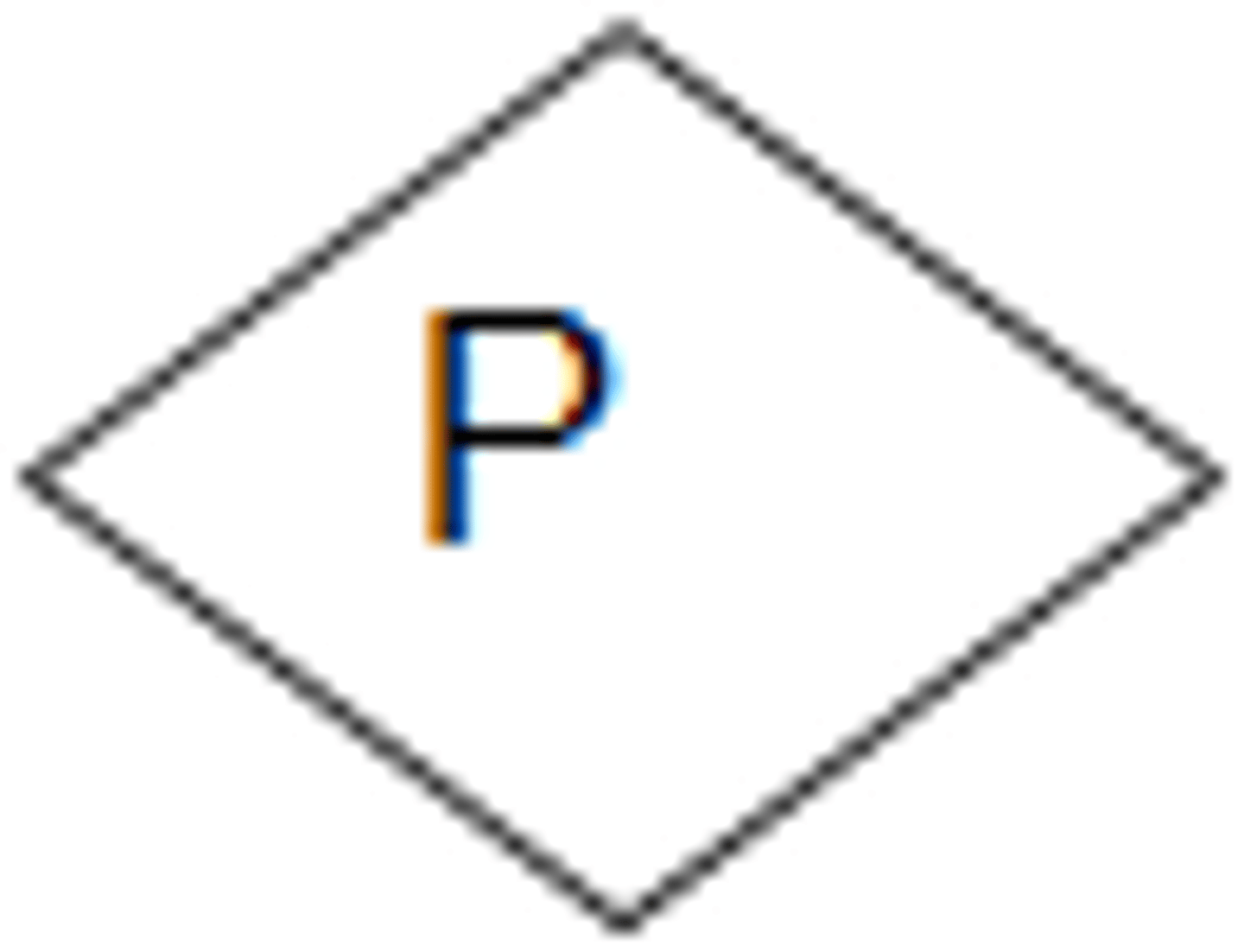
proband

Pregnancy
Ectopic pregnancy
Unaffected individual
Cannot have any alleles of a dominant trait (because a single allele of a dominant trait causes an individual to be affected)
Patterns that indicate a recessive trait
If any affected has 2 unaffected parents
If the trait is autosomal, both parents can be unaffected carriers of the disease
If the trait is x-linked, the son must have inherited his allele from his mother only, and his father can be unaffected
Autosomal recessive
If any affected founding daughter has 2 unaffected parents
Both parents must have an allele
A trait that skips a generation is a classic clue
May appear, disappear, and reappear across generations
Male/females equally affected
Unaffected mating having an affected offspring (both parents are heterozygous)
X-linked recessive
When an affected non-founding son has 2 unaffected parents the disease must be X-linked recessive
A male cannot be affected by a single autosomal recessive allele, but can be affected by a single X-linked recessive allele
More males than females affected, males need only one copy of the defective allele (hemizygous)
If no females (or only a few) are affected, it is likely that the trait is X-linked
Affected sons receive the allele on the X from their mothers
An affected female must have an affected father
Patterns that indicate a DOMINANT trait
The disease must be DOMINANT if every affected child of NON-FOUNDING parents has an affected parent.
No child could be affected by a single autosomal recessive allele, or X-linked recessive allele, so the trait is dominant.
Autosomal dominant
When affected son of non-founding parents has an affected father
A father does not transmit X-linked alleles to a son, so the disease cannot be X-linked dominant
When an affected daughter of non-founding parents has an affected father, we cannot determine whether the dominant disease is autosomal or x-linked. The father can transmit either an autosomal dominant allele, or an X-linked dominant allele to his daughter
Usually found in every generation
Males/females equally affected
An affected individual must have at least one affected parent
Affected individual usually has affected offspring
X-linked dominant
More females than males affected, females have 2 chances to receive allele (XX)
An affected individual must have at least one affected parent
Trait observed every generation (cannot skip a generation)
Cancer
Uncontrolled cell growth
Invasion
the spread of cancer cells from the original tumor to other parts of the body
Metastisis
the process by which cancer cells spread into nearby tissues
Angiogenesis
the process of new blood vessels forming from existing blood vessels
Green boxes
start codons