Chapter 3: Nucleic Acid Extraction Molecular Diagnostics Lela Buckingham
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
88 Terms
What is the purpose of Nucleic Acid Extraction
To release the nucleic acid from the cell for use in subsequent procedures. Makes sure the Nucleic acid is free from contamination with proteins, carbohydrates, lipids, DNA free of RNA and RNA free of DNA
What are some variables when it comes to DNA yield
WBC count (higher WBC in blood, more DNA), Type of specimen used and condition of specimen, cellularity, age, fixative, processing techniques: Digestion and fixation.
Blood Buffy coat (3.5-10^3 WBC/microliters) will yield how much DNA
50-100 micrograms
Bone Marrow (1 mL) will yield how much DNA
100-500 micrograms
Cultured Cells (10^7 cells) will yield how much DNA
30-70 micrograms
solid tissue (1 mg) will yield how much DNA
1-10 micrograms
Lavage fluids (10 mL) will yield how much DNA
2-250 micrograms
Mitochondria ( 10 mg tissue 10^7 cells) will yield how much DNA
1-10 micrograms
Plasmid DNA bacterial culture (100 mL overnight) will yield how much DNA
350 micrograms to 1 mg
Bacterial culture (0.5 mL 0.7 Absorbance units) will yield how much DNA
10-35 micrograms
Feces ( 1mg, bacteria or fungi) will yield how much DNA
2-228 micrograms
Serum/plasma, CSF (0.5 mL) will yield how much DNA
0.3-3 micrograms
Dried Blood (0.5-1cm diameter spot) will yield how much DNA
0.04-0.7 micrograms
Saliva (1 mL) will yield how much DNA
5-15 micrograms
Buccal cells (1 mg) will yield how much DNA
1-10 micrograms
Bone, teeth (500 mg) will yield how much DNA
30-50 micrograms
Hair Follicles will yield how much DNA
0.1-0.2 micrograms
Fixed tissues ( 5-10 10 micron sections) will yield how much DNA
6-50 micrograms
feces (animal cells, 1 mg) will yield how much DNA
2-100 picograms
Why do we isolate DNA with alkaline buffers
causes large chromosomal DNA and proteins (which we do not measure) to precipitate out of the solution
Bacteria and Fungi need their cell walls broken down to allow release of Nucleic acid before DNA/RNA can be analyzed. How is this done?
Enzyme products or mechanical breakdown. Commercial reagents, sucralose, Triton X-100 detergent, Tris buffer, EDTA after Lysozyme treatment releases DNA that can be precipitated with alcohol. Treatment with 1% SDS and 0.2M NaOH in Tris base/EDTA/Glucose can also do this.
Commercial reagents designed for isolation of DNA in amplification procedures like PCR are used for which organisms
Yeast, Fungi, and Gram positive Bacteria
What do Viruses Require for DNA isolation
Concentrations of cell free specimens (like plasma). This is to avoid contamination with other DNA/RNA
In Blood and Bone marrow aspirate, where is Nucleic acid mainly found?
Mainly in WBC. Blood is purified by differential density gradient centrifugation and Differential lysis to remove non-nucleic acid elements. Tghis is done by using Isotonic and Ficoll, centrifuging the product and taking out the Nucleic acid layer. You can also use osmotic fragility, Hypotonic saline to lyse the RBC then centrifuge to remove the RBC from WBC layer
In Differential density gradient centrifugation and Differential lysis, Which layer is the nucleic acid WBC in
The middle Layer. The top layer has RBC and PMN WBC->Nucleic acid containing WBC->layer of plasma
Solid tumors and transplanted organs release exosome cells and nucleic acid into the blood stream. What are exosome cells.
small vesicles formed by budding and invaginations from living cells; contain proteins, lipids and nucleic acids. Collected by centrifugation and be detected for diagnostic/ prognostic analysis
liquid biopsy
test done on a sample of blood to look for cancer cells, or pieces of DNA from tumor cells. Cancer sheds exosomes at a higher rate and can be quantified in this test
Nucleic acid quantification using tissue cells
Fresh/Frozen tissues must be dissociated before isolation. DNA in preserved tissues may be broken or crosslinked. DNA of 100 bp or less can be obtained from this specimen
Which tissue fixatives produce good quality Nucleic Acids
10% Buffered Neutral Formalin and Acetone (2.0-5.0 kb fragment size)
Which tissue fixatives produce moderate (not as good) quality Nucleic acids
Zambonis(0.2-2.0 kb fragment size), Clarkes(0.8-1.0 kb fragment size), Paraformaldehyde(0.2-5.0 kb fragment size), Metacam(0.7-1.5 kb fragment size), Formalin-Alcohol-Acetic Acid (1.0-4.0 kb fragment size)
Which tissue fixatives produce the worst quality (less Desirable) quality Nucleic acids
B-5(0.7-1.5 kb fragment size), Carnoys (0.7-1.5 kb fragment size), Bouins <0.1 kb
Organic isolation method DNA
1. Cells in suspension undergo lysis using NaOH and SDS detergent
2. acidification is performed using acidic acid and salt then vortex
3. Cell Debris is pelleted/extracted with chloroform
4. Biphasic emulsification occurs where the DNA in aq precipitate is in the top aq layer and the organic phase is in the lower layer
5. DNA is Centrifuged and precipitated with 1:1-2:1 ratio of ethanol: isopropanol alcohol
6. Excess salt is then removed by rinsing nucleic acid in760% ETOH, centrifuging, discarding supernatant and then dissolving DNA pellet in rehydration solution (10 mM Tris, 1 mM TE/EDTA or water)
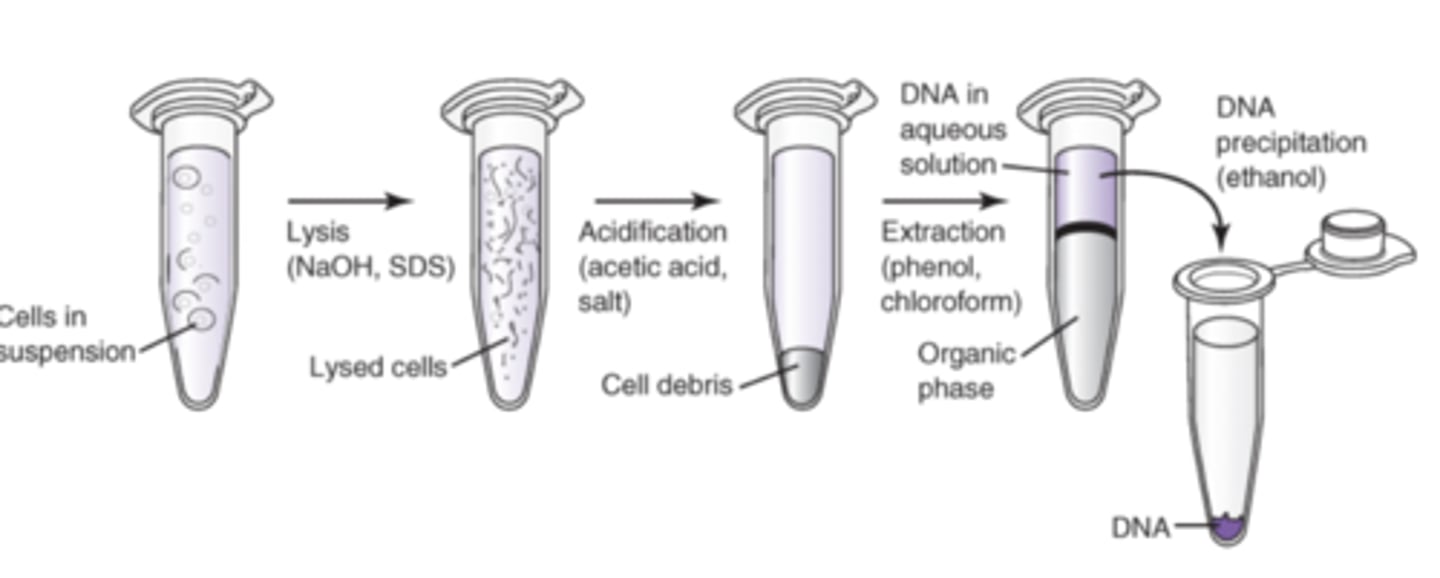
After DNA is released from the cell, it is still mixed and needs to be purified. How is this done?
Using High salt, Low pH and phenol and chloroform. Fungi is treated with cetyltrimethylammonium bromide- this helps separate DNA from polysaccharides. RNase is added to prevent RNA contamination. To form a solid precipitate of DNA add a 1:1-2:1 ratio of ethanol/isopropanol alcohol
Inorganic Isolation
1. Lyse using Tris Buffer, EDTA and SDS
2. Add low pH/high salt soln (Sodium acxetate to precipitate the proteins) > vortex/spin
3. Transfer aq soln layer to new tube (this has the DNA)
4. EtOH or isopropanol is used to precipitate DNA
5. Resuspend in a smaller volume of rehydration solution
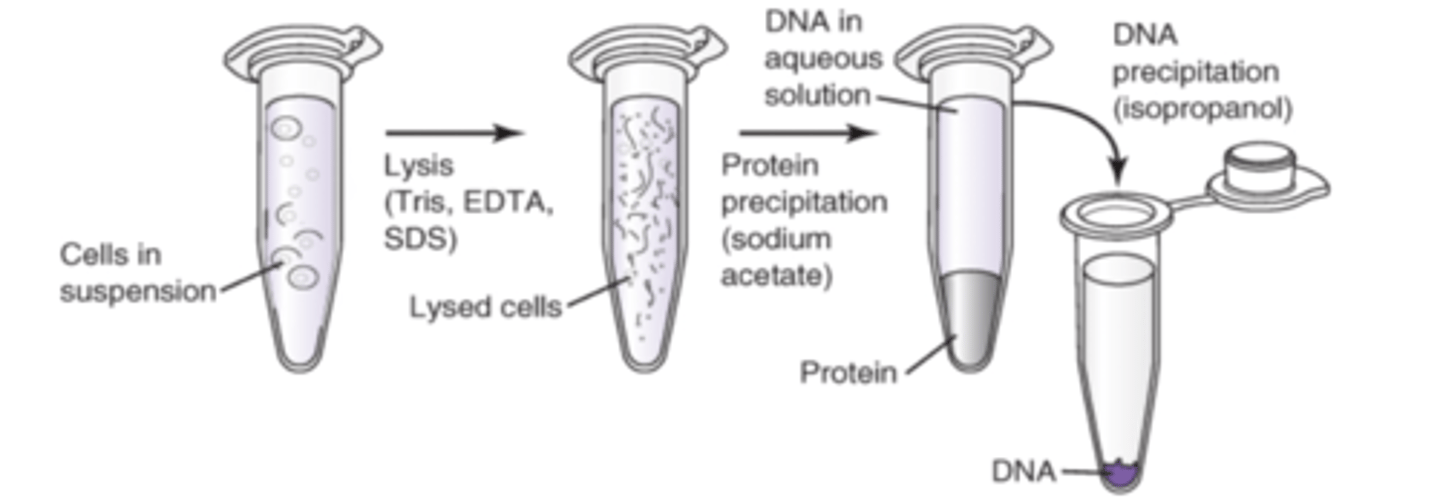
Concentration of DNA can be controlled by
adjusting buffer or water volume used for resuspension of the pellet. Don't dilute DNA so much you require a lot of EDTA/TE buffer
Why can alkaline lysis be used to specifically select plasmid DNA
Bc chromosomal DNA will not renature properly after neutralization & will form a precipitate nb
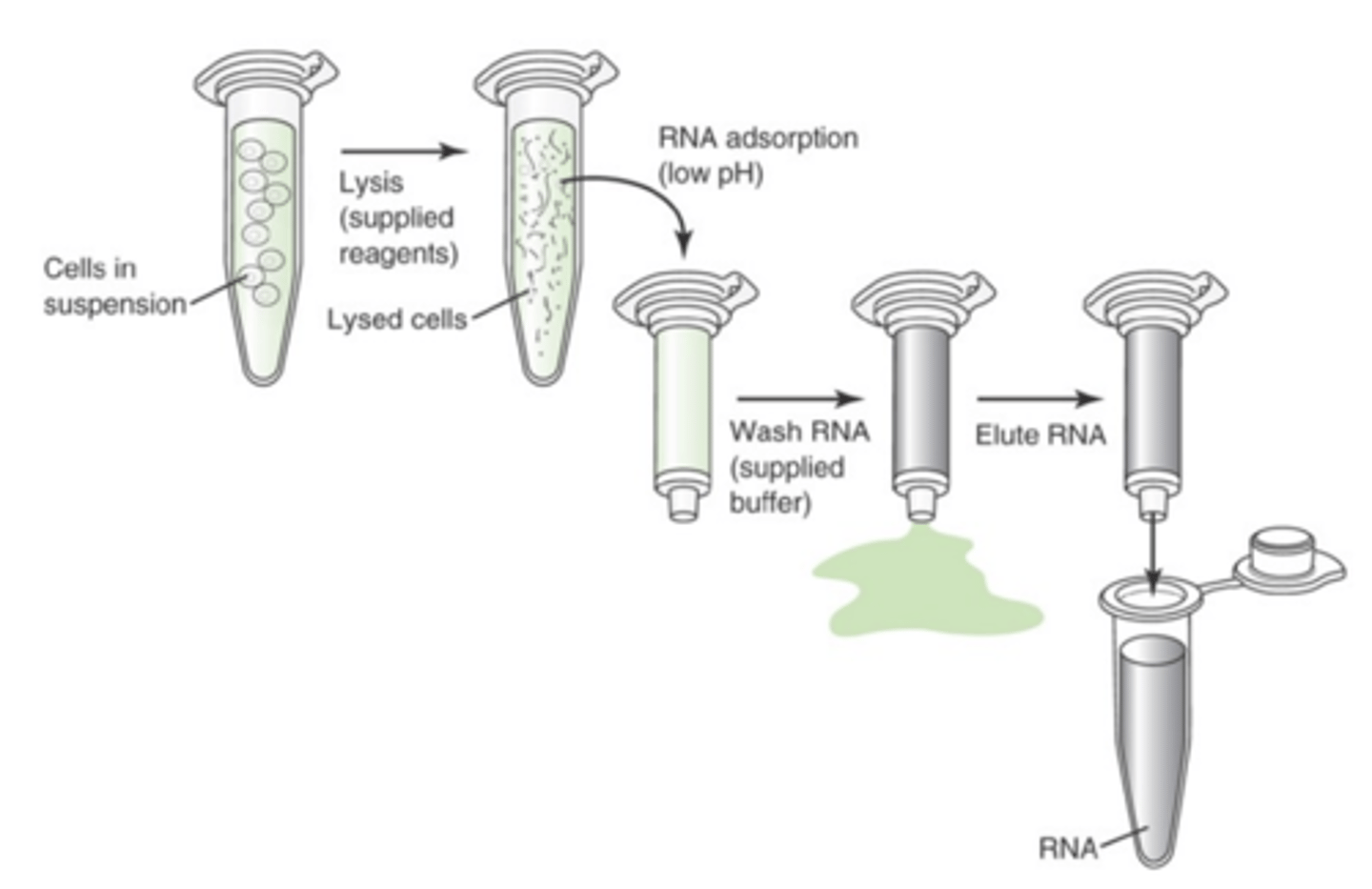
What are the steps in Solid phase isolation
1. Lyse (using supplied reagents)
2. Acidify (low pH using supplied reagents)
3. Transfer to column (adsorption)
4. Wash DNA using supplied buffer
5. Elute DNA using low salt
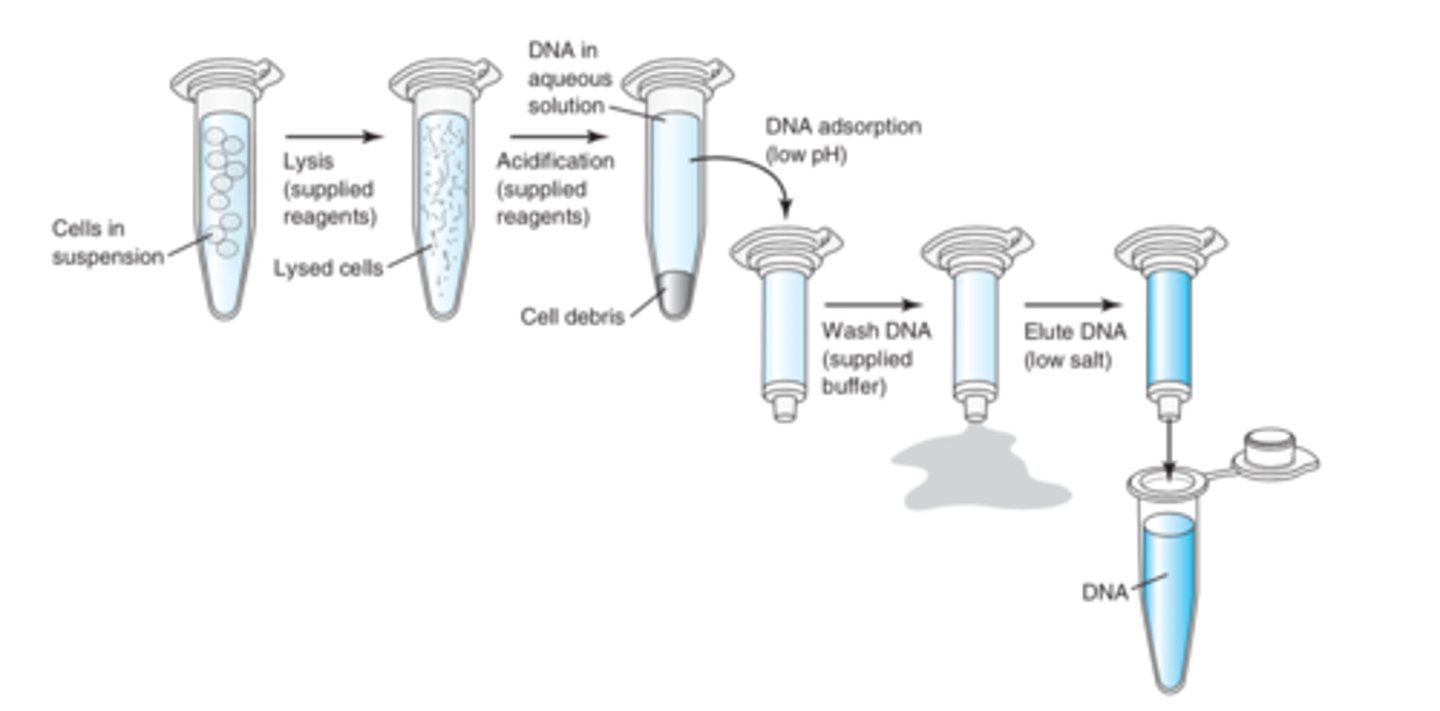
Describe solid phase isolation
more rapid and effective DNA extraction method. Uses solid matrixes to bind and hold DNA for washing. Methodologically employed by most robotic DNA isolation systems that used magnetized beads or membranes to bind DNA
Proteolytic lysis of fixed material steps
1. Tissue embedded in parafilm undergoes a microdissection to create 5-20 micron sections
2. Tissue is deparaffinized with a xylene and ETOH wash
3. Cellular material in the tissue is digested with proteinase K and Tris Buffer. This causes lysis of cellular material *IMP FOR DNA AMPLIFICATION*
Why would we perform Proteolytic lysis of fixed material
Sometimes you cannot purify all DNA and you need to screen large amounts of samples by simple methods, or isolate DNA from limited amounts of parafilm tissues given. This is used in research as a simple screening method when there are large amounts of samples that need to be processed
Describe the Rapid Extraction Method for DNA extraction
a Rapid Lysis method for DNA extraction that uses storage cards. Done by 1. 10% Chelix Resin beads are mixed with specimen and cells are lysed by boiling and then centrifuged 2. DNA in supernatant is cooled and extracted with chloroform before amplified. This is used in Forensics, Purification of DNA samples in paraffin embedded samples and DNA on spotted storage method
Isolation of mitochondria DNA
Done using rapid extraction methods and PCR/Hybridization or manually using the following way: 1. Cell prep is homogenized by grinding on ice (or using reducing-lysing agents to protect mitoDNA) then centrifuging on low speed 2.mitochrondria is pelleted in a high speed centrifuge, lysed with detergent, and then treated with proteinase to remove protein contaminants 3. mitoDNA is then precipitated with cold ETOH and resuspended in water or buffers
Isolation of RNA
extremely liable due to ubiquitous presence of RNases. Need to keep a RNAse free area of the lab.
RNA most to least abundant
80-90% of RNA is rRNA, mRNA is 205-5.0%, tRNA and snRNA makes up the remaining portion of RNA
Specimen collection for RNA
use special tubes/pellet RNA to stabilize RNA (can also add acid), Dimethyl Pyrocarbonate (DPEC) inactivates RNases
RNA isolation Chemistries
a little different than DNA. Cells are lysed by osmosis/separated from the WBC by centrifugation. When dissociating tissue, use liquid nitrogen/buffer that will inactivate intracellular RNases (same for bacteria and fungal RNA). Viral RNA is isolated using beads, must be cell free because test cannot differentiate RNA from host v microorganism.
RNA Organic Isolation
Cell Lysis step for RNA isolation is performed in detergent/phenol, in high salts/ RNase inhibitors. Acid phenol: chloroform: isoamyl alcohol alcohol (25:24:1) to Extract RNA.Treatment with DNAse to remove any contaminating DNA. Glycogen lyst transfer RNA may be added as a carrier to aid in RNA Pellet Formation
RNA Solid Phase Isolation
1 million eukaryotic 11-50 mg of tissue will yield 10 micrograms of RNA (depends on factors such as which type of tissue and paraffin.
Proteolytic lysis of fixed material
Fixation and handling of specimen before use will effect quality of RNA.
What are some factors that will affect the yield of RNA from a paraffin embedded tissue sample
Type of Fixative used, age/length of storage of the tissue, preliminary handling of the original specimen, time of fixation, type of specimen, specimen transport
Isolation of PolyA (messenger) RNA
2.5-5% of total RNA Yield. Mainly from highly expressed genes. To enrich yield of mRNA, ss oligomers of thymine and uracil is immobilized on a matrix of resin and beads. RNA is washed and polyA RNA is washed/ warmed with low salt buffer detergent to break H bonds between mRNA and column. 30-40 ng mRNA is obtained from 1 microgram of RNA. Limitations: efficiency is variable, the mixture is enriched with polyA RNA/not a pure PolyA prep.

Yield of RNA from Various specimen sources: Blood (1 mL 3.5-10 WBC/mL)
1-10 ug (Yield depends on WBC count)
Yield of RNA from Various specimen sources Buffy Coat (1 mL whole Blood)
5-10 ug (Yield depends on count)
Yield of RNA from Various specimen sources Bone Marrow (1 mL)
50-200 ug (Yield depends on cellularity)
Yield of RNA from Various specimen sources (Cultured cells 10^7 cells)
50-150 ug (Yield depends on type of cells)
Yield of RNA from Various specimen sources (Buccal Cells 1 mg)
1-10 ug
Yield of RNA from Various specimen sources (Solid Tissue)
0.5-4 ug ( Liver, spleen, heart yield more than Brain, lung, thymus, ovary, and kidney)
Yield of RNA from Various specimen sources (Fixed Tissue)
0.2-3 ug ( Yield depends on fixative used)
Yield of RNA from Various specimen sources (Bacterial Culture 0.5 mL 0.7 absorbance units)
10-100 ug (yield depends on type of bacterial culture grown)
Measurement of Nucleic Acid Quantity and Quality
Done by resolving an aliquot of a isolated sample on an agarose gel
RNA dyes
ethidium bromide or SybrGreen II
DNA dyes
Sybr Green I (main), Less frequently used: silver stain
What should RNA look like on an agarose gel
RNA is 2 bands 1 representing large and small RNA. If bands are smeared/degraded or absent, Quality of RNA is unacceptable
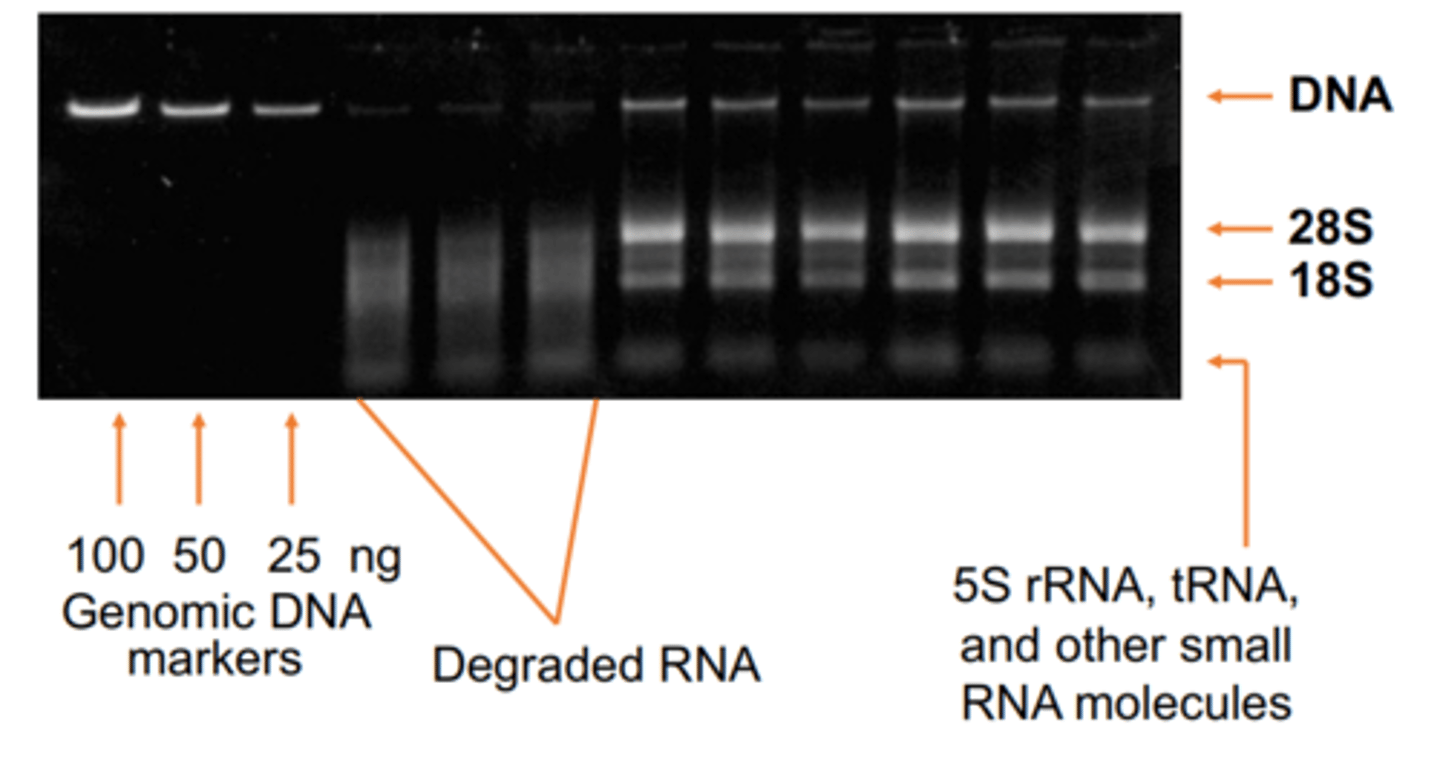
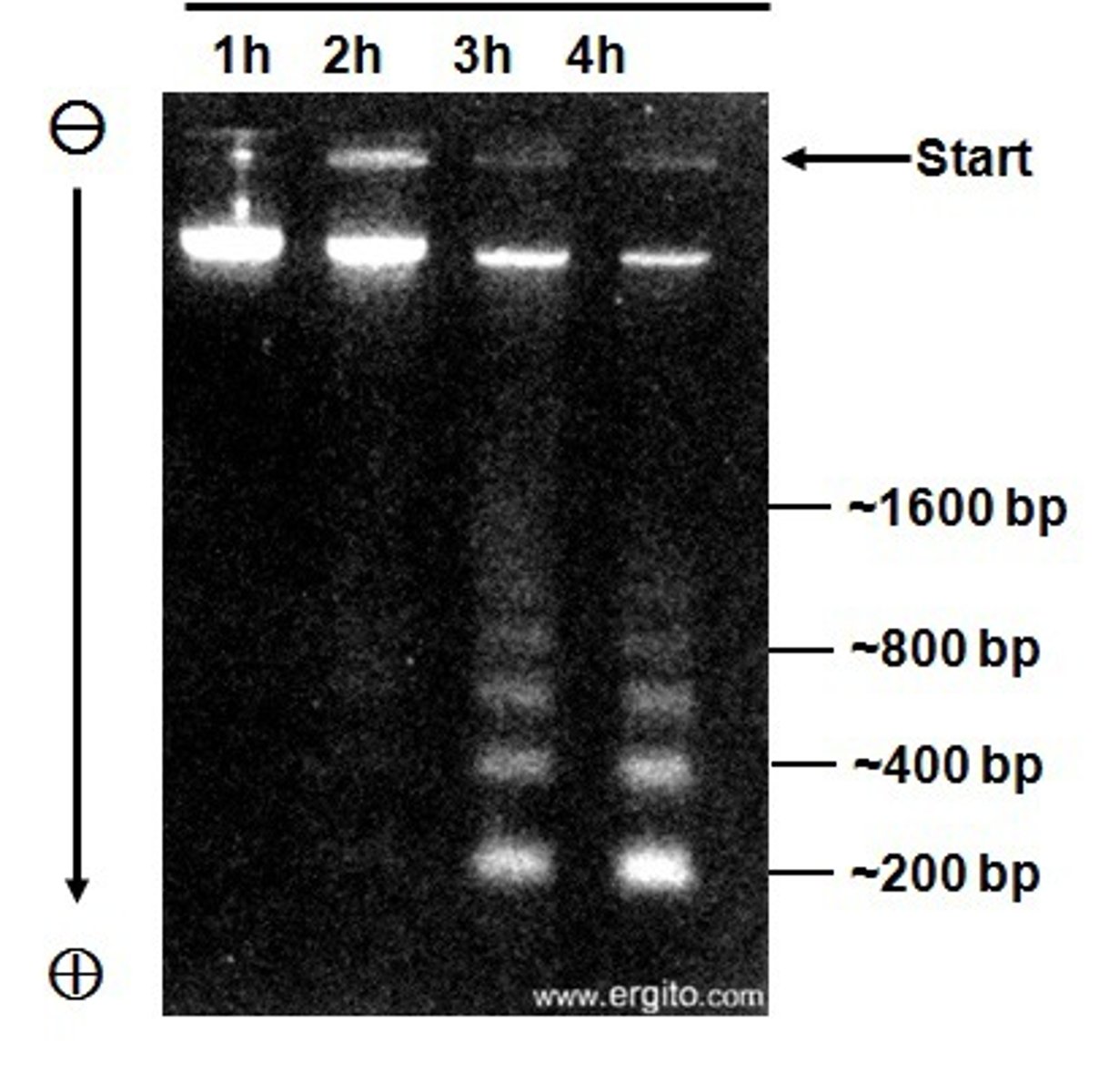
What should DNA look like on an agarose gel
Ideal DNA is a tight smear close to loading wells. Bright signal from supercoiled DNA. Minor/no signal from linear linked broken plasmids. High Molecular Weight chromosomal DNA collects at top.
Florescent dues can quantify by comparison of sample intensity with known DNA and control on same gel. How can this be done?
Densitometry of intensities of band is most quantitatively accurate and sometimes can be done visually.
When measuring the amount of DNA in a sample what do you have to multiply
50 ug/mL or mg/L
When measuring the amount of RNA in a sample what do you have to multiply
40 ug/mL or mg/L
Spectrophotometry
Nucleic acid absorbs light @ 260 nm through adenine residues. Beer-Lambert Law A=abc where A=Absorbance, a= molar absorptivity, b=path length, c=concentration (DNA 50, RNA 40). Spectrophotometric measurement may also be used to estimate purity of nucleic acid.
Common contaminants and their wavelengths of peak absorbance: Organic Compounds
230 nm
Common contaminants and their wavelengths of peak absorbance: Phenol
270 nm
Common contaminants and their wavelengths of peak absorbance: Protein
280 nm
Common contaminants and their wavelengths of peak absorbance: Particulate Matter
>330 nm
What is the most common contaminant
Protein
What should ideal DNA ratio absorbance be
Nucleic acid absorbance of 260 nm should be 1.6-2x more than absorbance at 280 nm. This makes sure the DNA is not too contaminated with protein. DNA with a absorbance higher than 2 maybe contaminated with RNA.
What should ideal RNA ratio absorbance be
Nucleic acid absorbance of 260 nm should be 2.0-2.3 more than absorbance at 280 nm.
Is spectrophotometry or Fluorometry considered a better method of Quality Analysis
Fluorometry is a better method of QA
Fluorometry
Measures Florescence related to DNA concentration in associated with DNA specific fluorescent dyes. Recommended for procedures where accurate measurement of intact DNA such as next generational sequence (NGS). More modern procedures use DNA specific Hoechst 33258, PicoGreen and OliGreen
Hoechst 33258 Dye
combines with AT base pairs in the minor groove of DNA--specific for double strand DNA and very sensitive (200 ng of DNA/mL)
Is Picogreen or Hoechst 33258 more sensitive
Picogreen 25pg/mL vs 200 pg/ML
OliGreen
Will bind to short single strand oligonucleotides of DNA (100 pg/mL of DNA). Will not fluoresce with ds DNA or RNA.
How is RNA measured with SybrGreen II
20-26% lower florescence with Polyadylated RNA compared to regular RNA. Measures as low as 2 ng/mL. Not specific to RNA, make sure that all DNA is out
How to calibrate Fluorometry
calibration with a known amount of standard before measurement of sample in a quartz cuvette within 2 hours
Absorbance and Fluorometry may not always agree. How?
Different targets, Single nucleotides don't bind to florescent dyes but they do absorb UV light and affect spectrophotometry. RNA doesn't enhance the florescence of the dyes and is invisible to fluorometric detection. Most labs use spectrophotometry
Microfluidics
Lab on chip has been applied to nucleic acid quantitation/Analysis using microfluidics based instruments. More automated, less sample, multiple tests can be done simultaneously
Compare and contrast measurement of DNA concentration with spectrophotometry and Florescence in regards to staining and accuracy
Spectrophotometry doesn't require staining but fluorometry does, Fluorometry is more accurate spectrophotometry can pick up spare nucleotides. Fluorescence does not decipher between DNA/RNA
After agarose gel electrophoresis, a 0.5 microgram aliquot of DNA isolated from a bacterial culture produced only a faint smear at the bottom of the gel lane. Is this an acceptable DNA sample?
This amount of DNA should produce a bright band near the top of the gel lane. This DNA is probs degraded and not good to use.
A blood sample was held at room temperature for 5 days before being processed for RNA isolation. Will this sample likely yield optimal RNA?
No. The RNA would degrade.
Name three factors that will affect the yield of RNA from a paraffin-embedded tissue sample
1.) Type of fixative used
2.) age/length of storage of the tissue
3.) preliminary handling of the original specimen
Also important: Time of fixation, type of specimen, specimen transport conditions.